3TTI
 
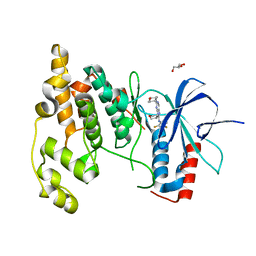 | | Crystal Structure of JNK3 complexed with CC-930, an orally active anti-fibrotic JNK inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 10, trans-4-({9-[(3S)-tetrahydrofuran-3-yl]-8-[(2,4,6-trifluorophenyl)amino]-9H-purin-2-yl}amino)cyclohexanol | | Authors: | Plantevin-Krenitsky, V, Nadolny, L, Delgado, M, Ayala, L, Clareen, S, Hilgraf, R, Albers, R, Hegde, S, D'Sidocky, N, Sapienza, J, Wright, J, McCarrick, M, Bahmanyar, S, Chamberlain, P, Delker, S.L, Muir, J, Giegel, D, Xu, L, Celeridad, M, Lachowitzer, J, Bennett, B, Moghaddam, M, Khatsenko, O, Katz, J, Fan, R, Bai, A, Tang, Y, Shirley, M.A, Benish, B, Bodine, T, Blease, K, Raymon, H, Cathers, B.E, Satoh, Y. | | Deposit date: | 2011-09-14 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of CC-930, an orally active anti-fibrotic JNK inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3TTJ
 
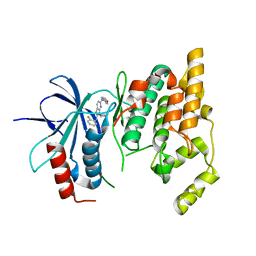 | | Crystal Structure of JNK3 complexed with CC-359, a JNK inhibitor for the prevention of ischemia-reperfusion injury | | Descriptor: | 9-cyclopentyl-N~8~-(2-fluorophenyl)-N~2~-(4-methoxyphenyl)-9H-purine-2,8-diamine, Mitogen-activated protein kinase 10 | | Authors: | Plantevin-Krenitsky, V, Delgado, M, Nadolny, L, Sahasrabudhe, K, Ayala, S, Clareen, S, Hilgraf, R, Albers, R, Kois, A, Hughes, K, Wright, J, Nowakowski, J, Sudbeck, E, Ghosh, S, Bahmanyar, S, Chamberlain, P, Muir, J, Cathers, B.E, Giegel, D, Xu, L, Celeridad, M, Moghaddam, M, Khatsenko, O, Omholt, P, Katz, J, Pai, S, Fan, R, Tang, Y, Shirley, M.A, Benish, B, Blease, K, Raymon, H, Bhagwat, S, Bennett, B, Satoh, Y. | | Deposit date: | 2011-09-14 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aminopurine based JNK inhibitors for the prevention of ischemia reperfusion injury.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2X8W
 
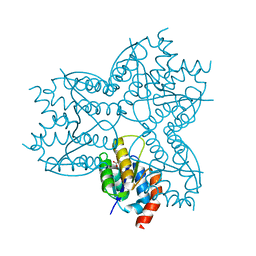 | | The Crystal Structure of Methylglyoxal Synthase from Thermus sp. GH5 Bound to Malonate. | | Descriptor: | MALONATE ION, METHYLGLYOXAL SYNTHASE | | Authors: | Shahsavar, A, Erfani Moghaddam, M, Antonyuk, S.V, Khajeh, K, Naderi-Manesh, H. | | Deposit date: | 2010-03-13 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Methylglyoxal Synthase from Thermus Sp.Gh5 in the Open and Closed Conformational States Provide Insight Into the Mechanism of Allosteric Regulation
To be Published
|
|
2XW6
 
 | | The Crystal Structure of Methylglyoxal Synthase from Thermus sp. GH5 Bound to Phosphate Ion. | | Descriptor: | METHYLGLYOXAL SYNTHASE, PHOSPHATE ION | | Authors: | Shahsavar, A, Erfani Moghaddam, M, Antonyuk, S.V, Khajeh, K, Naderi-Manesh, H. | | Deposit date: | 2010-11-01 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Atomic Resolution Structure of Methylglyoxal Synthase from Thermus Sp. Gh5 Bound to Phosphate: Insights Into the Distinctive Effects of Phosphate on the Enzyme Structure
To be Published
|
|
3QMX
 
 | |
