1BTC
 
 | | THREE-DIMENSIONAL STRUCTURE OF SOYBEAN BETA-AMYLASE DETERMINED AT 3.0 ANGSTROMS RESOLUTION: PRELIMINARY CHAIN TRACING OF THE COMPLEX WITH ALPHA-CYCLODEXTRIN | | Descriptor: | BETA-AMYLASE, BETA-MERCAPTOETHANOL, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Mikami, B, Hehre, E.J, Sato, M, Katsube, Y, Hirose, M, Morita, Y, Sacchettini, J.C. | | Deposit date: | 1993-02-18 | | Release date: | 1993-10-31 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0-A resolution structure of soybean beta-amylase complexed with alpha-cyclodextrin.
Biochemistry, 32, 1993
|
|
1BYA
 
 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
1BYB
 
 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
1BYC
 
 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
1BYD
 
 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION, alpha-D-glucopyranose-(1-4)-2-deoxy-beta-D-arabino-hexopyranose | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
4F13
 
 | | Alginate lyase A1-III Y246F complexed with tetrasaccharide | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-05-06 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F10
 
 | | Alginate lyase A1-III H192A complexed with tetrasaccharide | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-05-05 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4E1Y
 
 | | Alginate lyase A1-III H192A apo form | | Descriptor: | Alginate lyase | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-03-07 | | Release date: | 2012-04-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5EEO
 
 | | soybean lipoxygenase(L1)-T756R | | Descriptor: | FE (II) ION, Seed linoleate 13S-lipoxygenase-1 | | Authors: | Mikami, B, Nagaya, T, Hioki, Y. | | Deposit date: | 2015-10-23 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for specificity in lipoxygenase catalysis
To Be Published
|
|
1B90
 
 | | BACILLUS CEREUS BETA-AMYLASE APO FORM | | Descriptor: | ACETATE ION, CALCIUM ION, PROTEIN (BETA-AMYLASE), ... | | Authors: | Mikami, B, Adachi, M, Kage, T, Sarikaya, E, Nanmori, T, Shinke, R, Utsumi, S. | | Deposit date: | 1999-03-06 | | Release date: | 1999-03-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose.
Biochemistry, 38, 1999
|
|
1B9Z
 
 | | BACILLUS CEREUS BETA-AMYLASE COMPLEXED WITH MALTOSE | | Descriptor: | ACETATE ION, CALCIUM ION, PROTEIN (BETA-AMYLASE), ... | | Authors: | Mikami, B, Adachi, M, Kage, T, Sarikaya, E, Nanmori, T, Shinke, R, Utsumi, S. | | Deposit date: | 1999-03-06 | | Release date: | 1999-03-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose.
Biochemistry, 38, 1999
|
|
1B1Y
 
 | | SEVENFOLD MUTANT OF BARLEY BETA-AMYLASE | | Descriptor: | PROTEIN (BETA-AMYLASE), alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose | | Authors: | Mikami, B, Yoon, H.J, Yoshigi, N. | | Deposit date: | 1998-11-25 | | Release date: | 1998-12-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the sevenfold mutant of barley beta-amylase with increased thermostability at 2.5 A resolution.
J.Mol.Biol., 285, 1999
|
|
1C7G
 
 | |
2FH8
 
 | | Crystal Structure Analysis of Klebsiella pneumoniae pullulanase complexed with isomaltose | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-6)-beta-D-glucopyranose | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
2FHC
 
 | | Crystal Structure Analysis of Klebsiella pneumoniae pullulanase complexed with maltotriose | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
2FH6
 
 | | Crystal Structure Analysis of Klebsiella pneumoniae pullulanase complexed with glucose | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
2FHB
 
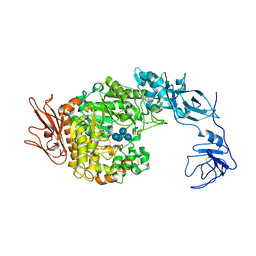 | | Crystal Structure Analysis of Klebsiella pneumoniae pullulanase complexed with maltose | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
2FGZ
 
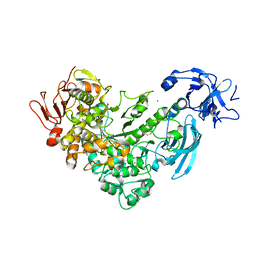 | | Crystal Structure Analysis of apo pullulanase from Klebsiella pneumoniae | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
2FHF
 
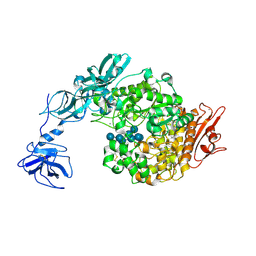 | | Crystal Structure Analysis of Klebsiella pneumoniae pullulanase complexed with maltotetraose | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
8ZRZ
 
 | |
5Y6Q
 
 | |
9JIP
 
 | | CYP105A1 R84A complexed with flufenamic acid (FLF) | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mikami, B, Takita, T, Sakaki, T, Yasuda, K, Wada, M, Yasukawa, K. | | Deposit date: | 2024-09-12 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure-Function Analysis of Streptomyces griseolus CYP105A1 in the Metabolism of Nonsteroidal Anti-inflammatory Drugs.
Biochemistry, 64, 2025
|
|
9JI6
 
 | | CYP105A1 R84A complexed with diclofenac (DIF) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, ... | | Authors: | Mikami, B, Takita, T, Sakaki, T, Yasuda, K, Wada, M, Yasukawa, K. | | Deposit date: | 2024-09-11 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Function Analysis of Streptomyces griseolus CYP105A1 in the Metabolism of Nonsteroidal Anti-inflammatory Drugs.
Biochemistry, 64, 2025
|
|
9JI1
 
 | | CYP105A1 complexed with flufenamic acid (FLF) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, ... | | Authors: | Mikami, B, Takita, T, Sakaki, T, Yasuda, K, Wada, M, Yasukawa, K. | | Deposit date: | 2024-09-11 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Function Analysis of Streptomyces griseolus CYP105A1 in the Metabolism of Nonsteroidal Anti-inflammatory Drugs.
Biochemistry, 64, 2025
|
|
9JHW
 
 | | CYP105A1 complexed with diclofenac (DIF) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mikami, B, Takita, T, Sakaki, T, Yasuda, K, Wada, M, Yasukawa, K. | | Deposit date: | 2024-09-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Function Analysis of Streptomyces griseolus CYP105A1 in the Metabolism of Nonsteroidal Anti-inflammatory Drugs.
Biochemistry, 64, 2025
|
|
