2LK5
 
 | | Solution structure of the Zn(II) form of Desulforedoxin | | Descriptor: | Desulforedoxin, ZINC ION | | Authors: | Goodfellow, B.J, Tavares, P, Romao, M.J, Czaja, C, Rusnak, F, Legall, J, Moura, I, Moura, J.J.G. | | Deposit date: | 2011-10-06 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of desulforedoxin, a simple iron-sulfur protein - An NMR study of the zinc derivative
J.BIOL.INORG.CHEM., 1, 1996
|
|
3KAQ
 
 | | Flavodoxin from D. desulfuricans (semireduced form) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Romero, A, Caldeira, J, LeGall, J, Moura, I, Moura, J.J.G, Romao, M.J. | | Deposit date: | 2009-10-19 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of flavodoxin from Desulfovibrio desulfuricans ATCC 27774 in two oxidation states.
Eur.J.Biochem., 239, 1996
|
|
3KAP
 
 | | Flavodoxin from Desulfovibrio desulfuricans ATCC 27774 (oxidized form) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Romero, A, Caldeira, J, LeGall, J, Moura, I, Moura, J.J.G, Romao, M.J. | | Deposit date: | 2009-10-19 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of flavodoxin from Desulfovibrio desulfuricans ATCC 27774 in two oxidation states.
Eur.J.Biochem., 239, 1996
|
|
1A2I
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO VULGARIS (HILDENBOROUGH) FERROCYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Kastrau, D.H.K, Costa, H.S, Legall, J, Turner, D.L, Santos, H, Xavier, A.V. | | Deposit date: | 1998-01-05 | | Release date: | 1998-07-08 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Desulfovibrio vulgaris (Hildenborough) ferrocytochrome c3: structural basis for functional cooperativity.
J.Mol.Biol., 281, 1998
|
|
1DVB
 
 | | RUBRERYTHRIN | | Descriptor: | FE (III) ION, RUBRERYTHRIN, ZINC ION | | Authors: | Sieker, L.C, Holmes, M, Le Trong, I, Turley, S, Liu, M.Y, Legall, J, Stenkamp, R.E. | | Deposit date: | 2000-01-20 | | Release date: | 2000-11-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A crystal structure of the "as isolated" rubrerythrin from Desulfovibrio vulgaris: some surprising results.
J.Biol.Inorg.Chem., 5, 2000
|
|
1SPW
 
 | | Solution Structure of a Loop Truncated Mutant from D. gigas Rubredoxin, NMR | | Descriptor: | Rubredoxin | | Authors: | Pais, T.M, Lamosa, P, dos Santos, W, LeGall, J, Turner, D.L, Santos, H. | | Deposit date: | 2004-03-17 | | Release date: | 2005-03-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of protein stabilization by solutes: the importance of the hairpin loop in rubredoxins
FEBS J., 272, 2005
|
|
1GYO
 
 | | Crystal structure of the di-tetraheme cytochrome c3 from Desulfovibrio gigas at 1.2 Angstrom resolution | | Descriptor: | CYTOCHROME C3, A DIMERIC CLASS III C-TYPE CYTOCHROME, GLYCEROL, ... | | Authors: | Aragao, D, Frazao, C, Sieker, L, Sheldrick, G.M, Legall, J, Carrondo, M.A. | | Deposit date: | 2002-04-29 | | Release date: | 2002-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Dimeric Cytochrome C3 from Desulfovibrio Gigas at 1.2 A Resolution
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NF6
 
 | | X-ray structure of the Desulfovibrio desulfuricans bacterioferritin: the diiron site in different catalytic states ("cycled" structure: reduced in solution and allowed to reoxidise before crystallisation) | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, FE (III) ION, GLYCEROL, ... | | Authors: | Macedo, S, Romao, C.V, Mitchell, E, Matias, P.M, Liu, M.Y, Xavier, A.V, LeGall, J, Teixeira, M, Lindley, P, Carrondo, M.A. | | Deposit date: | 2002-12-13 | | Release date: | 2003-04-01 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The nature of the di-iron site in the bacterioferritin from
Desulfovibrio desulfuricans
NAT.STRUCT.BIOL., 10, 2003
|
|
1NF4
 
 | | X-Ray Structure of the Desulfovibrio desulfuricans bacterioferritin: the diiron site in different states (reduced structure) | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, FE (II) ION, SULFATE ION, ... | | Authors: | Macedo, S, Romao, C.V, Mitchell, E, Matias, P.M, Liu, M.Y, Xavier, A.V, LeGall, J, Teixeira, M, Lindley, P, Carrondo, M.A. | | Deposit date: | 2002-12-13 | | Release date: | 2003-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The nature of the di-iron site in the bacterioferritin from
Desulfovibrio desulfuricans
NAT.STRUCT.BIOL., 10, 2003
|
|
1NFV
 
 | | X-ray structure of Desulfovibrio desulfuricans bacterioferritin: the diiron centre in different catalytic states (as-isolated structure) | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, 3-HYDROXYPYRUVIC ACID, FE (III) ION, ... | | Authors: | Macedo, S, Romao, C.V, Mitchell, E, Matias, P.M, Liu, M.Y, Xavier, A.V, LeGall, J, Teixeira, M, Lindley, P, Carrondo, M.A. | | Deposit date: | 2002-12-16 | | Release date: | 2003-04-01 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The nature of the di-iron site in the bacterioferritin from
Desulfovibrio desulfuricans
NAT.STRUCT.BIOL., 10, 2003
|
|
1QN1
 
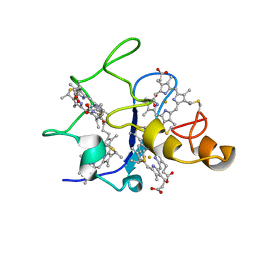 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERRICYTOCHROME C3, NMR, 15 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Brennan, L, Messias, A.C, Legall, J, Turner, D.L, Xavier, A.V. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-12 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Network of Functional Cooperativities in Cytochromes C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
1QN0
 
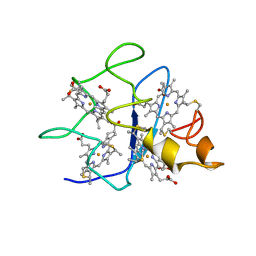 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERROCYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Teodoro, M.L, Brennan, L, Legall, J, Santos, H, Xavier, A.V, Turner, D.L. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-12 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Network of Functional Cooperativities in Cytochrome C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
1H21
 
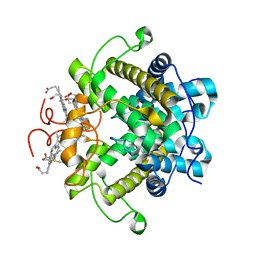 | | A novel iron centre in the split-Soret cytochrome c from Desulfovibrio desulfuricans ATCC 27774 | | Descriptor: | HEME C, SPLIT-SORET CYTOCHROME C | | Authors: | Abreu, I.A, Lourenco, A.I, Xavier, A.V, Legall, J, Coelho, A.V, Matias, P.M, Pinto, D.M, Carrondo, M.A, Teixeira, M, Saraiva, L.M. | | Deposit date: | 2002-07-30 | | Release date: | 2003-02-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Iron Centre in the Split-Soret Cytochrome C from Desulfovibrio Desulfuricans Atcc 27774
J.Biol.Inorg.Chem., 8, 2003
|
|
1KCB
 
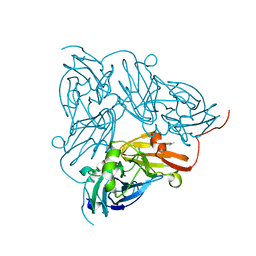 | | Crystal Structure of a NO-forming Nitrite Reductase Mutant: an Analog of a Transition State in Enzymatic Reaction | | Descriptor: | COPPER (II) ION, Nitrite Reductase | | Authors: | Liu, S.Q, Chang, T, Liu, M.Y, LeGall, J, Chang, W.C, Zhang, J.P, Liang, D.C, Chang, W.R. | | Deposit date: | 2001-11-07 | | Release date: | 2003-11-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a NO-forming nitrite reductase mutant: an analog of a transition state in enzymatic reaction
Biochem.Biophys.Res.Commun., 302, 2003
|
|
1OA1
 
 | | REDUCED HYBRID CLUSTER PROTEIN (HCP) FROM DESULFOVIBRIO VULGARIS HILDENBOROUGH STRUCTURE AT 1.55A RESOLUTION USING SYNCHROTRON RADIATION. | | Descriptor: | FE4-S3 CLUSTER, GLYCEROL, HYDROXYLAMINE REDUCTASE, ... | | Authors: | Aragao, D, Macedo, S, Mitchell, E.P, Romao, C.V, Liu, M.Y, Frazao, C, Saraiva, L.M, Xavier, A.V, Legall, J, Van Dongen, W.M.A.M, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P.F. | | Deposit date: | 2002-12-23 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Reduced Hybrid Cluster Proteins (Hcp) from Desulfovibrio Desulfuricans Atcc 27774 and Desulfovibrio Vulgaris (Hildenborough): X-Ray Structures at High Resolution Using Synchrotron Radiation
J.Biol.Inorg.Chem., 8, 2003
|
|
1OA0
 
 | | REDUCED HYBRID CLUSTER PROTEIN FROM DESULFOVIBRIO DESULFURICANS X-RAY STRUCTURE AT 1.25A RESOLUTION | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FE4-S3 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Macedo, S, Aragao, D, Mitchell, E.P, Romao, C.V, Liu, M.Y, Frazao, C, Saraiva, L.M, Xavier, A.V, Legall, J, Van Dongen, W.M.A.M, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P.F. | | Deposit date: | 2002-12-23 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Reduced hybrid cluster proteins (HCP) from Desulfovibrio desulfuricans ATCC 27774 and Desulfovibrio vulgaris (Hildenborough): X-ray structures at high resolution using synchrotron radiation.
J. Biol. Inorg. Chem., 8, 2003
|
|
1GNT
 
 | | Hybrid Cluster Protein from Desulfovibrio vulgaris. X-ray structure at 1.25A resolution using synchrotron radiation. | | Descriptor: | HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, IRON/SULFUR/OXYGEN HYBRID CLUSTER | | Authors: | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hybrid cluster proteins (HCPs) from Desulfovibrio desulfuricans ATCC 27774 and Desulfovibrio vulgaris (Hildenborough): X-ray structures at 1.25 A resolution using synchrotron radiation.
J. Biol. Inorg. Chem., 7, 2002
|
|
1B71
 
 | | RUBRERYTHRIN | | Descriptor: | FE (III) ION, PROTEIN (RUBRERYTHRIN), ZINC ION | | Authors: | Sieker, L.C, Holmes, M, Le Trong, I, Turley, S, Santarsiero, B.D, Liu, M.-Y, Legall, J, Stenkamp, R.E. | | Deposit date: | 1999-01-26 | | Release date: | 2000-01-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternative metal-binding sites in rubrerythrin.
Nat.Struct.Biol., 6, 1999
|
|
1H29
 
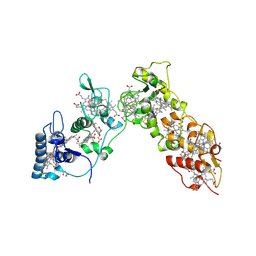 | | Sulfate respiration in Desulfovibrio vulgaris Hildenborough: Structure of the 16-heme Cytochrome c HmcA at 2.5 A resolution and a view of its role in transmembrane electron transfer | | Descriptor: | HEME C, HIGH-MOLECULAR-WEIGHT CYTOCHROME C | | Authors: | Matias, P.M, Coelho, A.V, Valente, F.M.A, Placido, D, Legall, J, Xavier, A.V, Pereira, I.A.C, Carrondo, M.A. | | Deposit date: | 2002-08-01 | | Release date: | 2002-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Sulfate Respiration in Desulfovibrio Vulgaris Hildenborough: Structure of the 16-Heme Cytochrome C Hmca at 2.5 A Resolution and a View of its Role in Transmembrane Electron Transfer
J.Biol.Chem., 277, 2002
|
|
1GN9
 
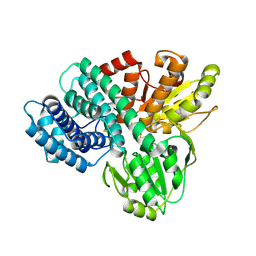 | | Hybrid Cluster Protein from Desulfovibrio desulfuricans ATCC 27774 X-ray structure at 2.6A resolution using synchrotron radiation at a wavelength of 1.722A | | Descriptor: | HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, IRON/SULFUR/OXYGEN HYBRID CLUSTER | | Authors: | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | Deposit date: | 2001-10-04 | | Release date: | 2002-04-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hybrid Cluster Proteins (Hcps) from Desulfovibrio Desulfuricans Atcc 27774 and Desulfovibrio Vulgaris (Hildenborough): X-Ray Structures at 1.25 A Resolution Using Synchrotron Radiation.
J.Biol.Inorg.Chem., 7, 2002
|
|
1GNL
 
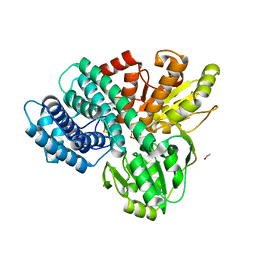 | | Hybrid Cluster Protein from Desulfovibrio desulfuricans X-ray structure at 1.25A resolution using synchrotron radiation at a wavelength of 0.933A | | Descriptor: | ACETATE ION, HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | Deposit date: | 2001-10-05 | | Release date: | 2002-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hybrid Cluster Proteins (Hcps) from Desulfovibrio Desulfuricans Atcc 27774 and Desulfovibrio Vulgaris (Hildenborough): X-Ray Structures at 1.25 A Resolution Using Synchrotron Radiation.
J.Biol.Inorg.Chem., 7, 2002
|
|
2NRD
 
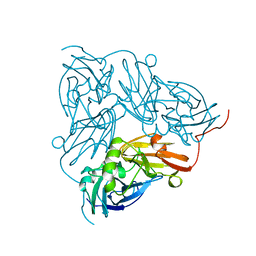 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
6RXN
 
 | |
1JYB
 
 | | Crystal structure of Rubrerythrin | | Descriptor: | FE (III) ION, Rubrerythrin, ZINC ION | | Authors: | Chang, W.R, Li, M, Liu, M.Y. | | Deposit date: | 2001-09-11 | | Release date: | 2002-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure studies on rubrerythrin: enzymatic activity in relation to the zinc movement.
J.Biol.Inorg.Chem., 8, 2003
|
|
1NIF
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
