5SVH
 
 | |
2LOV
 
 | | AR55 solubilised in LPPG micelles | | Descriptor: | Apelin receptor | | Authors: | Langelaan, D.N, Rainey, J.K. | | Deposit date: | 2012-01-27 | | Release date: | 2013-01-16 | | Last modified: | 2023-07-26 | | Method: | SOLUTION NMR | | Cite: | Preserved Transmembrane Segment Topology, Structure, and Dynamics in Disparate Micellar Environments.
J Phys Chem Lett, 8, 2017
|
|
2LOW
 
 | | Solution structure of AR55 in 50% HFIP | | Descriptor: | Apelin receptor | | Authors: | Langelaan, D.N, Rainey, J.K. | | Deposit date: | 2012-01-27 | | Release date: | 2013-01-16 | | Last modified: | 2023-07-26 | | Method: | SOLUTION NMR | | Cite: | Preserved Transmembrane Segment Topology, Structure, and Dynamics in Disparate Micellar Environments.
J Phys Chem Lett, 8, 2017
|
|
5IBW
 
 | | Complex of MlcC bound to the tandem IQ motif of MyoC | | Descriptor: | Calcium-binding EF-hand domain-containing protein, Myosin IC heavy chain, SODIUM ION | | Authors: | Langelaan, D.N, Smith, S.P. | | Deposit date: | 2016-02-22 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Single-lobe Myosin Light Chain C in Complex with the Light Chain-binding Domains of Myosin-1C Provides Insights into Divergent IQ Motif Recognition.
J.Biol.Chem., 291, 2016
|
|
2LOU
 
 | | AR55 solubilised in DPC micelles | | Descriptor: | Apelin receptor | | Authors: | Langelaan, D.N, Rainey, J.K. | | Deposit date: | 2012-01-27 | | Release date: | 2013-01-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural features of the apelin receptor N-terminal tail and first transmembrane segment implicated in ligand binding and receptor trafficking.
Biochim.Biophys.Acta, 1828, 2013
|
|
2LOT
 
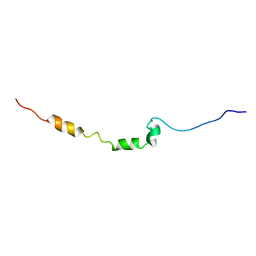 | | AR55 solubilised in SDS micelles | | Descriptor: | Apelin receptor | | Authors: | Langelaan, D.N, Rainey, J.K. | | Deposit date: | 2012-01-27 | | Release date: | 2013-01-16 | | Last modified: | 2023-07-26 | | Method: | SOLUTION NMR | | Cite: | Preserved Transmembrane Segment Topology, Structure, and Dynamics in Disparate Micellar Environments.
J Phys Chem Lett, 8, 2017
|
|
8E1D
 
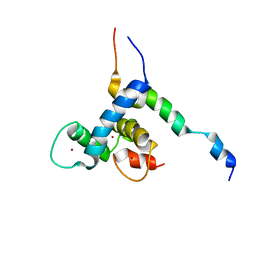 | |
2MH0
 
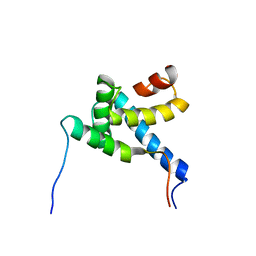 | |
7S86
 
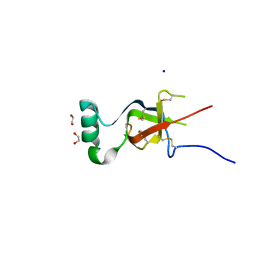 | |
7S7S
 
 | |
5W0Y
 
 | |
6E9M
 
 | |
6E98
 
 | |
2M8U
 
 | | Solution structure of the Dictyostelium discodieum Myosin Light Chain, MlcC | | Descriptor: | Myosin Light Chain, MlcC | | Authors: | Liburd, J.D, Miller, E, Langelaan, D, Chitayat, S, Crawley, S.W, Cote, G.P, Smith, S.P. | | Deposit date: | 2013-05-28 | | Release date: | 2014-12-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the Single-lobe Myosin Light Chain C in Complex with the Light Chain-binding Domains of Myosin-1C Provides Insights into Divergent IQ Motif Recognition.
J.Biol.Chem., 291, 2016
|
|
5IX9
 
 | | Cell surface anchoring domain | | Descriptor: | Antifreeze protein | | Authors: | Guo, S, Langelaan, D. | | Deposit date: | 2016-03-23 | | Release date: | 2017-06-28 | | Last modified: | 2020-01-08 | | Method: | SOLUTION NMR | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
5JUH
 
 | | Crystal structure of C-terminal domain (RV) of MpAFP | | Descriptor: | Antifreeze protein, CALCIUM ION | | Authors: | Guo, S. | | Deposit date: | 2016-05-10 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
5K8G
 
 | |
5T7A
 
 | | Crystal structure of Br derivative BhCBM56 | | Descriptor: | 1,2-ETHANEDIOL, BH0236 protein, BROMIDE ION | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-02 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Properties of a family 56 carbohydrate-binding module and its role in the recognition and hydrolysis of beta-1,3-glucan.
J. Biol. Chem., 292, 2017
|
|
2NBH
 
 | |
5IRB
 
 | | Structural insight into host cell surface retention of a 1.5-MDa bacterial ice-binding adhesin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Guo, S, Phippen, S, Campbell, R, Davies, P. | | Deposit date: | 2016-03-12 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
5J6Y
 
 | | Crystal structure of PA14 domain of MpAFP Antifreeze protein | | Descriptor: | Antifreeze protein, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Guo, S. | | Deposit date: | 2016-04-05 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
