1DIV
 
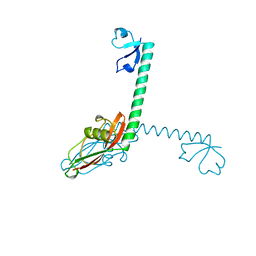 | | RIBOSOMAL PROTEIN L9 | | Descriptor: | RIBOSOMAL PROTEIN L9 | | Authors: | Hoffman, D.W, Cameron, C, Davies, C, Gerchman, S.E, Kycia, J.H, Porter, S, Ramakrishnan, V, White, S.W. | | Deposit date: | 1996-07-02 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of prokaryotic ribosomal protein L9: a bi-lobed RNA-binding protein.
EMBO J., 13, 1994
|
|
1Q3B
 
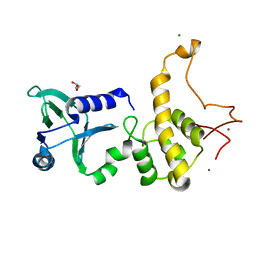 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli: The R252A mutant at 2.05 resolution. | | Descriptor: | Endonuclease VIII, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Golan, G, Zharkov, D.O, Feinberg, H, Fernandes, A.S, Zaika, E.I, Kycia, J.H, Grollman, A.P, Shoham, G. | | Deposit date: | 2003-07-29 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the uncomplexed DNA repair enzyme endonuclease VIII indicates significant interdomain flexibility.
Nucleic Acids Res., 33, 2005
|
|
1Q39
 
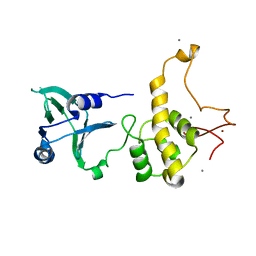 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli: The WT enzyme at 2.8 resolution. | | Descriptor: | CALCIUM ION, Endonuclease VIII, ZINC ION | | Authors: | Golan, G, Zharkov, D.O, Feinberg, H, Fernandes, A.S, Zaika, E.I, Kycia, J.H, Grollman, A.P, Shoham, G. | | Deposit date: | 2003-07-29 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the uncomplexed DNA repair enzyme endonuclease VIII indicates significant interdomain flexibility.
Nucleic Acids Res., 33, 2005
|
|
1Q3C
 
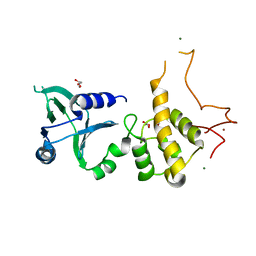 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli: The E2A mutant at 2.3 resolution. | | Descriptor: | Endonuclease VIII, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Golan, G, Zharkov, D.O, Feinberg, H, Fernandes, A.S, Zaika, E.I, Kycia, J.H, Grollman, A.P, Shoham, G. | | Deposit date: | 2003-07-29 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the uncomplexed DNA repair enzyme endonuclease VIII indicates significant interdomain flexibility.
Nucleic Acids Res., 33, 2005
|
|
1KG5
 
 | | Crystal structure of the K142Q mutant of E.coli MutY (core fragment) | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1KG6
 
 | | Crystal structure of the K142R mutant of E.coli MutY (core fragment) | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1KG4
 
 | | Crystal structure of the K142A mutant of E. coli MutY (core fragment) | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1KG3
 
 | | Crystal structure of the core fragment of MutY from E.coli at 1.55A resolution | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1KG2
 
 | | Crystal structure of the core fragment of MutY from E.coli at 1.2A resolution | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1KG7
 
 | | Crystal Structure of the E161A mutant of E.coli MutY (core fragment) | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1K3X
 
 | | Crystal structure of a trapped reaction intermediate of the DNA repair enzyme Endonuclease VIII with Brominated-DNA | | Descriptor: | 5'-D(*CP*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*(BRU)P*(BRU)P*CP*AP*(BRU)P*CP*CP*(BRU)P*GP*G)-3', Endonuclease VIII, ... | | Authors: | Golan, G, Zharkov, D.O, Gilboa, R, Fernandes, A.S, Kycia, J.H, Gerchman, S.E, Rieger, R.A, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate.
EMBO J., 21, 2002
|
|
1K82
 
 | | Crystal structure of E.coli formamidopyrimidine-DNA glycosylase (Fpg) covalently trapped with DNA | | Descriptor: | 5'-D(*CP*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*TP*TP*CP*CP*TP*CP*CP*TP*GP*G)-3', ZINC ION, ... | | Authors: | Gilboa, R, Zharkov, D.O, Golan, G, Fernandes, A.S, Gerchman, S.E, Matz, E, Kycia, J.H, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-10-22 | | Release date: | 2002-06-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA.
J.Biol.Chem., 277, 2002
|
|
1K3W
 
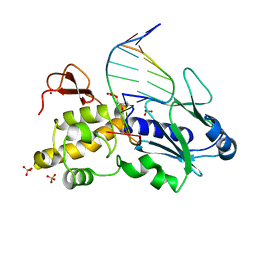 | | Crystal structure of a trapped reaction intermediate of the DNA Repair Enzyme Endonuclease VIII with DNA | | Descriptor: | 5'-D(*CP*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*TP*TP*CP*AP*TP*CP*CP*TP*GP*G)-3', Endonuclease VIII, ... | | Authors: | Golan, G, Zharkov, D.O, Gilboa, R, Fernandes, A.S, Kycia, J.H, Gerchman, S.E, Rieger, R.A, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate.
EMBO J., 21, 2002
|
|
1WHI
 
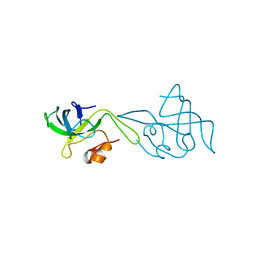 | | RIBOSOMAL PROTEIN L14 | | Descriptor: | RIBOSOMAL PROTEIN L14 | | Authors: | Davies, C, White, S.W, Ramakrishnan, V. | | Deposit date: | 1996-01-10 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of ribosomal protein L14 reveals an important organizational component of the translational apparatus.
Structure, 4, 1996
|
|
1JZT
 
 | |
1TIG
 
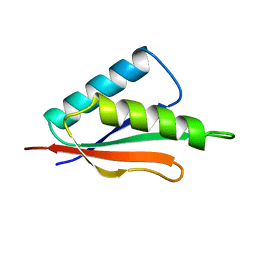 | | TRANSLATION INITIATION FACTOR 3 C-TERMINAL DOMAIN | | Descriptor: | TRANSLATION INITIATION FACTOR 3 | | Authors: | Biou, V, Shu, F, Ramakrishnan, V. | | Deposit date: | 1995-08-16 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallography shows that translational initiation factor IF3 consists of two compact alpha/beta domains linked by an alpha-helix.
EMBO J., 14, 1995
|
|
1TIF
 
 | | TRANSLATION INITIATION FACTOR 3 N-TERMINAL DOMAIN | | Descriptor: | CHLORIDE ION, TRANSLATION INITIATION FACTOR 3, TRIMETHYL LEAD ION | | Authors: | Biou, V, Shu, F, Ramakrishnan, V. | | Deposit date: | 1995-08-16 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallography shows that translational initiation factor IF3 consists of two compact alpha/beta domains linked by an alpha-helix.
EMBO J., 14, 1995
|
|
1CQU
 
 | |
