3BOY
 
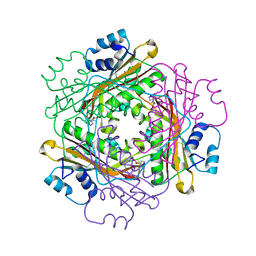 | | Crystal structure of the HutP antitermination complex bound to the HUT mRNA | | Descriptor: | 5'-R(*UP*UP*UP*AP*GP*UP*UP*UP*UP*UP*AP*GP*UP*UP*UP*UP*UP*AP*GP*UP*UP*U)-3', HISTIDINE, Hut operon positive regulatory protein, ... | | Authors: | Kumarevel, T.S, Balasundaresan, D, Jeyakanthan, J, Shinkai, A, Yokoyama, S, Kumar, P.K.R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-12-18 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of HutP complexed with the 55-mer RNA
To be Published
|
|
1NBK
 
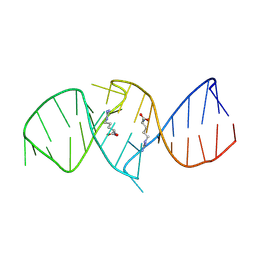 | | The structure of RNA aptamer for HIV Tat complexed with two argininamide molecules | | Descriptor: | 2-AMINO-5-GUANIDINO-PENTANOIC ACID, RNA aptamer | | Authors: | Matsugami, A, Kobayashi, S, Ouhashi, K, Uesugi, S, Yamamoto, R, Taira, K, Nishikawa, S, Kumar, P.K.R, Katahira, M. | | Deposit date: | 2002-12-03 | | Release date: | 2003-12-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Highly Efficient Trapping of the HIV Tat Protein by an RNA Aptamer
Structure, 11, 2003
|
|
1WRQ
 
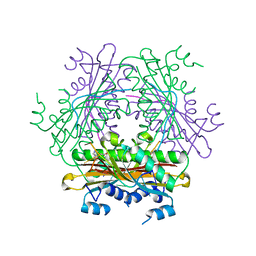 | |
1WRO
 
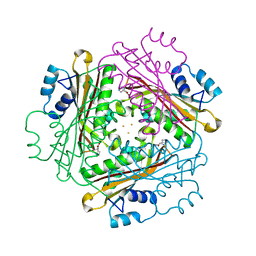 | | Metal Ion dependency of the antiterminator protein, HutP, for binding to the terminator region of hut mRNA- A structural basis | | Descriptor: | BARIUM ION, HISTIDINE, Hut operon positive regulatory protein | | Authors: | Kumarevel, T, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2004-10-25 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Characterization of the metal ion binding site in the anti-terminator protein, HutP, of Bacillus subtilis
Nucleic Acids Res., 33, 2005
|
|
1WRN
 
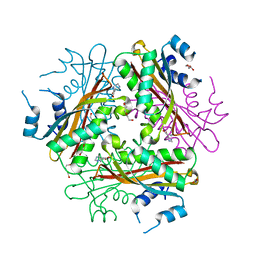 | | Metal Ion dependency of the antiterminator protein, HutP, for binding to the terminator region of hut mRNA- A structural basis | | Descriptor: | DI(HYDROXYETHYL)ETHER, HISTIDINE, Hut operon positive regulatory protein, ... | | Authors: | Kumarevel, T, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2004-10-25 | | Release date: | 2005-08-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the metal ion binding site in the anti-terminator protein, HutP, of Bacillus subtilis
Nucleic Acids Res., 33, 2005
|
|
4H4L
 
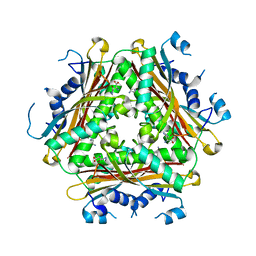 | | Crystal Structure of ternary complex of HutP(HutP-L-His-Zn) | | Descriptor: | HISTIDINE, Hut operon positive regulatory protein, ZINC ION | | Authors: | Dhakshnamoorthy, B, Misono, T.S, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2012-09-17 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Alternative binding modes of l-histidine guided by metal ions for the activation of the antiterminator protein HutP of Bacillus subtilis.
J.Struct.Biol., 183, 2013
|
|
1WPT
 
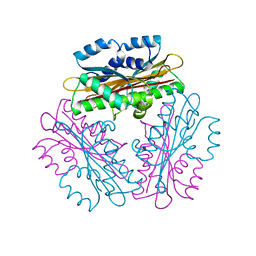 | | Crystal Structure of HutP, an RNA binding anti-termination protein | | Descriptor: | Hut operon positive regulatory protein | | Authors: | Kumarevel, T, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2004-09-13 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterization of the metal ion binding site in the anti-terminator protein, HutP, of Bacillus subtilis
Nucleic Acids Res., 33, 2005
|
|
1WPU
 
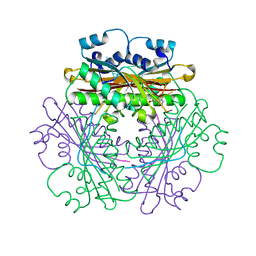 | |
2ZFA
 
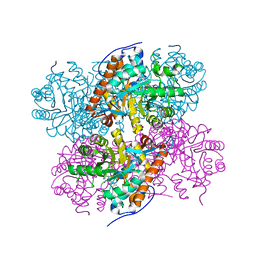 | | Structure of Lactate Oxidase at pH4.5 from AEROCOCCUS VIRIDANS | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Lactate oxidase | | Authors: | Furuichi, M, Balasundaresan, D, Suzuki, N, Yoshida, Y, Minagawa, H, Kaneko, H, Waga, I, Kumar, P.K.R, Mizuno, H. | | Deposit date: | 2007-12-26 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray structures of Aerococcus viridans lactate oxidase and its complex with D-lactate at pH 4.5 show an alpha-hydroxyacid oxidation mechanism
J.Mol.Biol., 378, 2008
|
|
1WMQ
 
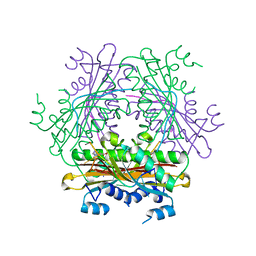 | | Structure of the HutP antitermination complex bound to a single stranded region of hut mRNA | | Descriptor: | 5'-R(P*UP*UP*UP*AP*GP*UP*U)-3', HISTIDINE, Hut operon positive regulatory protein, ... | | Authors: | Kumarevel, T.S, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2004-07-14 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of HutP-mediated anti-termination and roles of the Mg2+ ion and L-histidine ligand.
Nature, 434, 2005
|
|
1WPV
 
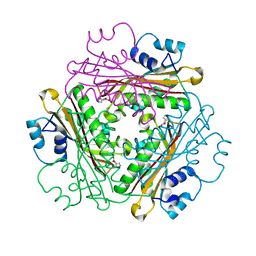 | | Crystal Structure of Activated Binary complex of HutP, an RNA binding anti-termination protein | | Descriptor: | HISTIDINE, Hut operon positive regulatory protein, MAGNESIUM ION | | Authors: | Kumarevel, T.S, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2004-09-14 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of HutP-mediated anti-termination and roles of the Mg2+ ion and L-histidine ligand.
Nature, 434, 2005
|
|
1VEA
 
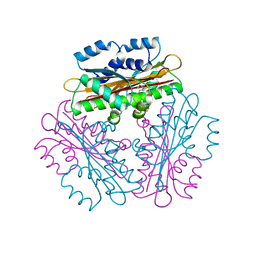 | | Crystal Structure of HutP, an RNA binding antitermination protein | | Descriptor: | Hut operon positive regulatory protein, N-(2-NAPHTHYL)HISTIDINAMIDE | | Authors: | Kumarevel, T.S, Fujimoto, Z, Karthe, P, Oda, M, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2004-03-29 | | Release date: | 2004-07-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Activated HutP; An RNA Binding Protein that Regulates Transcription of the hut Operon in Bacillus subtilis
Structure, 12, 2004
|
|
1WPS
 
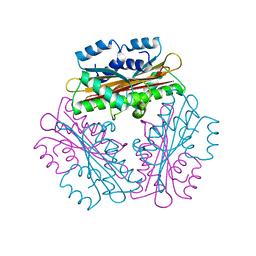 | |
2NLI
 
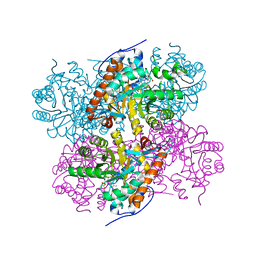 | | Crystal Structure of the complex between L-lactate oxidase and a substrate analogue at 1.59 angstrom resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, HYDROGEN PEROXIDE, LACTIC ACID, ... | | Authors: | Furuichi, M, Suzuki, N, Balasundaresan, D, Yoshida, Y, Minagawa, H, Watanabe, Y, Kaneko, H, Waga, I, Kumar, P.K.R, Mizuno, H. | | Deposit date: | 2006-10-20 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-ray structures of Aerococcus viridans lactate oxidase and its complex with D-lactate at pH 4.5 show an alpha-hydroxyacid oxidation mechanism
J.Mol.Biol., 378, 2008
|
|
2YX4
 
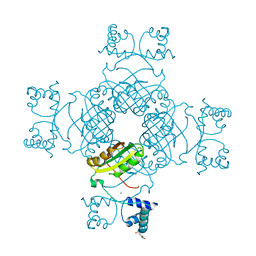 | | Crystal Structure of T134A of ST1022 from Sulfolobus tokodaii | | Descriptor: | 150aa long hypothetical transcriptional regulator, GLUTAMINE, MAGNESIUM ION | | Authors: | Kumarevel, T.S, Karthe, P, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-24 | | Release date: | 2008-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding
Nucleic Acids Res., 36, 2008
|
|
2EB7
 
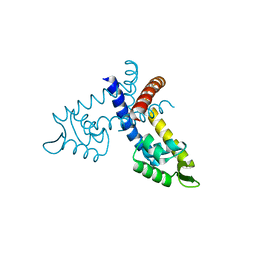 | | Crystal structure of the hypothetical regulator from Sulfolobus tokodaii 7 | | Descriptor: | 146aa long hypothetical transcriptional regulator | | Authors: | Kumarevel, T.S, Nishio, M, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-07 | | Release date: | 2007-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the MarR family regulatory protein, ST1710, from Sulfolobus tokodaii strain 7
J.Struct.Biol., 161, 2008
|
|
2PMH
 
 | | Crystal structure of Thr132Ala of ST1022 from Sulfolobus tokodaii | | Descriptor: | 150aa long hypothetical transcriptional regulator, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Kumarevel, T.S, Karthe, P, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-22 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding
Nucleic Acids Res., 36, 2008
|
|
