1W7W
 
 | |
1EHW
 
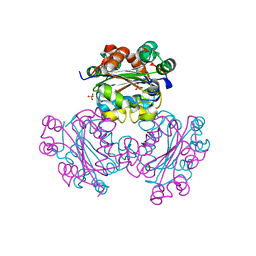 | | HUMAN NUCLEOSIDE DIPHOSPHATE KINASE 4 | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE, SULFATE ION | | Authors: | Milon, L, Meyer, P, Chiadmi, M, Munier, A, Johansson, M, Karlsson, A, Lascu, I, Capeau, J, Janin, J, Lacombe, M.-L. | | Deposit date: | 2000-02-23 | | Release date: | 2000-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The human nm23-H4 gene product is a mitochondrial nucleoside diphosphate kinase.
J.Biol.Chem., 275, 2000
|
|
5A7U
 
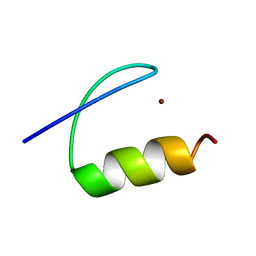 | | Single-particle cryo-EM of co-translational folded adr1 domain inside the E. coli ribosome exit tunnel. | | Descriptor: | REGULATORY PROTEIN ADR1, ZINC ION | | Authors: | Nilsson, O.B, Hedman, R, Marino, J, Wickles, S, Bischoff, L, Johansson, M, Muller-Lucks, A, Trovato, F, Puglisi, J.D, O'Brien, E, Beckmann, R, von Heijne, G. | | Deposit date: | 2015-07-10 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cotranslational Protein Folding Inside the Ribosome Exit Tunnel.
Cell Rep., 12, 2015
|
|
1GJT
 
 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN G | | Authors: | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | Deposit date: | 2001-08-02 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
1GJS
 
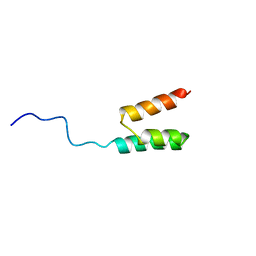 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN G | | Authors: | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | Deposit date: | 2001-08-02 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
6YRO
 
 | | Streptococcus suis SadP mutant - N285D | | Descriptor: | GLYCEROL, SODIUM ION, SadP | | Authors: | Papageorgiou, A.C, Haataja, S. | | Deposit date: | 2020-04-20 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The binding mechanism of the virulence factor Streptococcus suis adhesin P subtype to globotetraosylceramide is associated with systemic disease.
J.Biol.Chem., 295, 2020
|
|
2M4Q
 
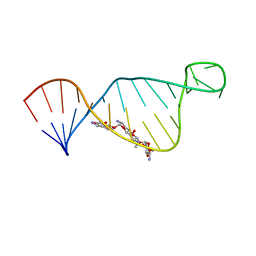 | | NMR structure of E. coli ribosomela decoding site with apramycin | | Descriptor: | APRAMYCIN, RNA (27-MER) | | Authors: | Puglisi, J.D, Tsai, A, Marshall, R, Viani, E. | | Deposit date: | 2013-02-10 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The impact of aminoglycosides on the dynamics of translation elongation.
Cell Rep, 3, 2013
|
|
