2ANW
 
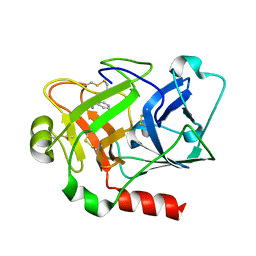 | | Expression, crystallization and three-dimensional structure of the catalytic domain of human plasma kallikrein: Implications for structure-based design of protease inhibitors | | Descriptor: | BENZAMIDINE, plasma kallikrein, light chain | | Authors: | Tang, J, Yu, C.L, Williams, S.R, Springman, E, Jeffery, D, Sprengeler, P.A, Estevez, A, Sampang, J, Shrader, W, Spencer, J.R, Young, W.B, McGrath, M.E, Katz, B.A. | | Deposit date: | 2005-08-11 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Expression, crystallization, and three-dimensional structure of the catalytic domain of human plasma kallikrein.
J.Biol.Chem., 280, 2005
|
|
2ANY
 
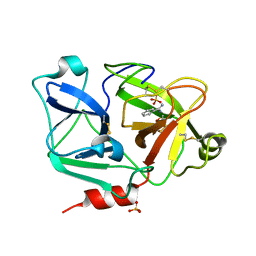 | | Expression, Crystallization and the Three-dimensional Structure of the Catalytic Domain of Human Plasma Kallikrein: Implications for Structure-Based Design of Protease Inhibitors | | Descriptor: | BENZAMIDINE, PHOSPHATE ION, plasma kallikrein, ... | | Authors: | Tang, J, Yu, C.L, Williams, S.R, Springman, E, Jeffery, D, Sprengeler, P.A, Estevez, A, Sampang, J, Shrader, W, Spencer, J.R, Young, W.B, McGrath, M.E, Katz, B.A. | | Deposit date: | 2005-08-11 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Expression, crystallization, and three-dimensional structure of the catalytic domain of human plasma kallikrein.
J.Biol.Chem., 280, 2005
|
|
5FUF
 
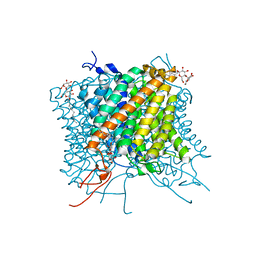 | | Crystal structure of the Mep2 mutant S453D from Candida albicans | | Descriptor: | AMMONIUM TRANSPORTER, DECYL-BETA-D-MALTOPYRANOSIDE | | Authors: | van den Berg, B, Chembath, A, Jefferies, D, Basle, A, Khalid, S, Rutherford, J. | | Deposit date: | 2016-01-26 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.066 Å) | | Cite: | Structural Basis for Mep2 Ammonium Transceptor Activation by Phosphorylation.
Nat.Commun., 7, 2016
|
|
5AEZ
 
 | |
5AEX
 
 | |
5AID
 
 | |
5AF1
 
 | |
5AH3
 
 | |
