4INK
 
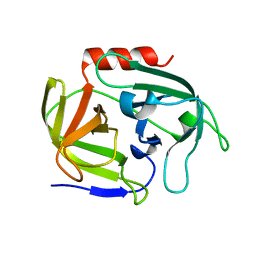 | | Crystal structure of SplD protease from Staphylococcus aureus at 1.56 A resolution | | Descriptor: | Serine protease SplD | | Authors: | Zdzalik, M, Kalinska, M, Cichon, P, Wysocka, M, Stec-Niemczyk, J, Stennicke, H.R, Jabaiah, A, Markiewicz, M, Wladyka, B, Daugherty, P.S, Lesner, A, Rolka, K, Dubin, A, Potempa, J, Dubin, G. | | Deposit date: | 2013-01-04 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Biochemical and Structural Characterization of SplD Protease from Staphylococcus aureus.
Plos One, 8, 2013
|
|
4INL
 
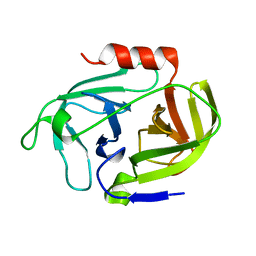 | | Crystal structure of SplD protease from Staphylococcus aureus at 2.1 A resolution | | Descriptor: | Serine protease SplD | | Authors: | Cichon, P, Zdzalik, M, Kalinska, M, Wysocka, M, Stec-Niemczyk, J, Stennicke, H.R, Jabaiah, A, Markiewicz, M, Wladyka, B, Daugherty, P.S, Lesner, A, Rolka, K, Dubin, A, Potempa, J, Dubin, G. | | Deposit date: | 2013-01-04 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and Structural Characterization of SplD Protease from Staphylococcus aureus.
Plos One, 8, 2013
|
|
5MM8
 
 | |
2XS4
 
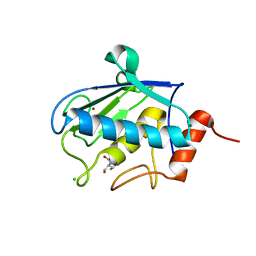 | | Structure of karilysin catalytic MMP domain in complex with magnesium | | Descriptor: | CHLORIDE ION, KARILYSIN PROTEASE, MAGNESIUM ION, ... | | Authors: | Cerda-Costa, N, Guevara, T, Karim, A.Y, Ksiazek, M, Nguyen, K.-A, Arolas, J.L, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of the Catalytic Domain of Tannerella Forsythia Karilysin Reveals It is a Bacterial Xenologue of Animal Matrix Metalloproteinases.
Mol.Microbiol., 79, 2011
|
|
2XS3
 
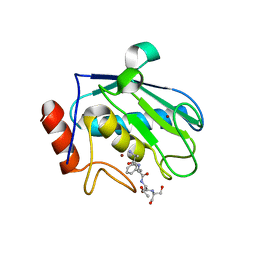 | | Structure of karilysin catalytic MMP domain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, KARILYSIN PROTEASE, PEPTIDE ALA-PHE-THR-SER, ... | | Authors: | Cerda-Costa, N, Guevara, T, Karim, A.Y, Ksiazek, M, Nguyen, K.-A, Arolas, J.L, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of the Catalytic Domain of Tannerella Forsythia Karilysin Reveals It is a Bacterial Xenologue of Animal Matrix Metalloproteinases.
Mol.Microbiol., 79, 2011
|
|
