2ZJD
 
 | | Crystal Structure of LC3-p62 complex | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B precursor, undecameric peptide from Sequestosome-1 | | Authors: | Ichimura, Y, Kumanomidou, T, Sou, Y, Mizushima, T, Ezaki, J, Ueno, T, Kominami, E, Yamane, T, Tanaka, K, Komatsu, M. | | Deposit date: | 2008-03-05 | | Release date: | 2008-06-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural Basis for Sorting Mechanism of p62 in Selective Autophagy
J.Biol.Chem., 283, 2008
|
|
3WIM
 
 | | GABARAP-LIR peptide complex | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, WD repeat and FYVE domain-containing protein 3 | | Authors: | Lystad, A, Ichimura, Y, Takagi, K, Yang, Y, Pankiv, S, Mizushima, T, Komatsu, M, Simonsen, A. | | Deposit date: | 2013-09-19 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | GABARAP-LIR peptide complex
To be Published, 2014
|
|
3WDZ
 
 | | Crystal Structure of Keap1 in Complex with phosphorylated p62 | | Descriptor: | Kelch-like ECH-associated protein 1, Peptide from Sequestosome-1 | | Authors: | Fukutomi, T, Takagi, K, Mizushima, T, Tanaka, K, Komatsu, M, Yamamoto, M. | | Deposit date: | 2013-06-26 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy.
Mol.Cell, 51, 2013
|
|
2DYT
 
 | |
4ZY3
 
 | | Crystal Structure of Keap1 in Complex with a small chemical compound, K67 | | Descriptor: | FORMIC ACID, Kelch-like ECH-associated protein 1, N,N'-[2-(2-oxopropyl)naphthalene-1,4-diyl]bis(4-ethoxybenzenesulfonamide) | | Authors: | Fukutomi, T, Iso, T, Suzuki, T, Takagi, K, Mizushima, T, Komatsu, M, Yamamoto, M. | | Deposit date: | 2015-05-21 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming
Nat Commun, 7, 2016
|
|
3ADE
 
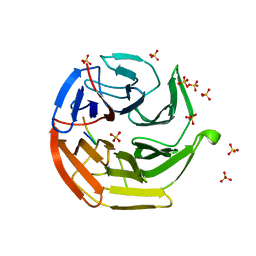 | | Crystal Structure of Keap1 in Complex with Sequestosome-1/p62 | | Descriptor: | Kelch-like ECH-associated protein 1, SULFATE ION, Sequestosome-1 | | Authors: | Kurokawa, H, Yamamoto, M. | | Deposit date: | 2010-01-19 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1
Nat.Cell Biol., 12, 2010
|
|
5HKH
 
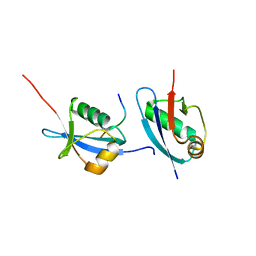 | | Crystal structure of Ufm1 in complex with UBA5 | | Descriptor: | ASP-ASN-GLU-TRP-GLY-ILE-GLU-LEU-VAL, Ubiquitin-fold modifier 1 | | Authors: | Huber, J, Doetsch, V, Rogov, V.V, Akutsu, M. | | Deposit date: | 2016-01-14 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Functional Analysis of a Novel Interaction Motif within UFM1-activating Enzyme 5 (UBA5) Required for Binding to Ubiquitin-like Proteins and Ufmylation.
J.Biol.Chem., 291, 2016
|
|
