2IO5
 
 | | Crystal structure of the CIA- histone H3-H4 complex | | Descriptor: | ASF1A protein, Histone H3.1, Histone H4 | | Authors: | Natsume, R, Akai, Y, Horikoshi, M, Senda, T. | | Deposit date: | 2006-10-10 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4.
Nature, 446, 2007
|
|
2RNZ
 
 | | Solution structure of the presumed chromodomain of the yeast histone acetyltransferase, Esa1 | | Descriptor: | Histone acetyltransferase ESA1 | | Authors: | Shimojo, H, Sano, N, Moriwaki, Y, Okuda, M, Horikoshi, M, Nishimura, Y. | | Deposit date: | 2008-03-01 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Novel structural and functional mode of a knot essential for RNA binding activity of the Esa1 presumed chromodomain
J.Mol.Biol., 378, 2008
|
|
2RO0
 
 | | Solution structure of the knotted tudor domain of the yeast histone acetyltransferase, Esa1 | | Descriptor: | Histone acetyltransferase ESA1 | | Authors: | Shimojo, H, Sano, N, Moriwaki, Y, Okuda, M, Horikoshi, M, Nishimura, Y. | | Deposit date: | 2008-03-01 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Novel structural and functional mode of a knot essential for RNA binding activity of the Esa1 presumed chromodomain
J.Mol.Biol., 378, 2008
|
|
1IMJ
 
 | |
1IXV
 
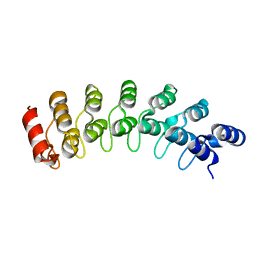 | | Crystal Structure Analysis of homolog of oncoprotein gankyrin, an interactor of Rb and CDK4/6 | | Descriptor: | Probable 26S proteasome regulatory subunit p28 | | Authors: | Padmanabhan, B, Adachi, N, Kataoka, K, Horikoshi, M. | | Deposit date: | 2002-07-09 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the homolog of the oncoprotein gankyrin, an interactor of Rb and CDK4/6
J.BIOL.CHEM., 279, 2004
|
|
1KP0
 
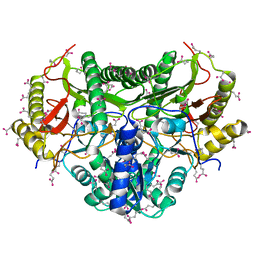 | |
2E50
 
 | | Crystal structure of SET/TAF-1beta/INHAT | | Descriptor: | Protein SET, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Muto, S, Senda, M, Senda, T, Horikoshi, M. | | Deposit date: | 2006-12-18 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Relationship between the structure of SET/TAF-Ibeta/INHAT and its histone chaperone activity
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3AAD
 
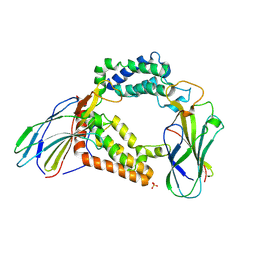 | | Structure of the histone chaperone CIA/ASF1-double bromodomain complex linking histone modifications and site-specific histone eviction | | Descriptor: | Histone chaperone ASF1A, SULFATE ION, Transcription initiation factor TFIID subunit 1 | | Authors: | Akai, Y, Adachi, N, Hayashi, Y, Eitoku, M, Sano, N, Natsume, R, Kudo, N, Tanokura, M, Senda, T, Horikoshi, M. | | Deposit date: | 2009-11-16 | | Release date: | 2010-04-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the histone chaperone CIA/ASF1-double bromodomain complex linking histone modifications and site-specific histone eviction
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2Z8U
 
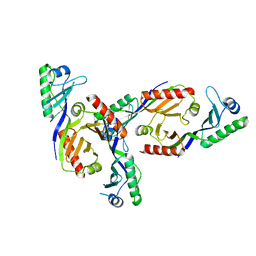 | | Methanococcus jannaschii TBP | | Descriptor: | TATA-box-binding protein | | Authors: | Adachi, N, Senda, T, Horikoshi, M. | | Deposit date: | 2007-09-10 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Methanococcus jannaschii TATA box-binding protein.
Genes Cells, 13, 2008
|
|
1WG0
 
 | | Structural comparison of Nas6p protein structures in two different crystal forms | | Descriptor: | Probable 26S proteasome regulatory subunit p28 | | Authors: | Nakamura, Y, Umehara, T, Tanaka, A, Horikoshi, M, Yokoyama, S, Padmanabhan, B, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2005-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural comparison of Nas6p protein structures in two different crystal forms
To be Published
|
|
1WG3
 
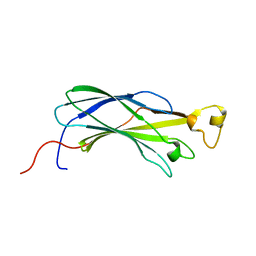 | | Structural analysis of yeast nucleosome-assembly factor CIA1p | | Descriptor: | Anti-silencing protein 1 | | Authors: | Padmanabhan, B, Kataoka, K, Umehara, T, Adachi, N, Yokoyama, S, Horikoshi, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Similarity between Histone Chaperone Cia1p/Asf1p and DNA-Binding Protein NF-{kappa}B
J.Biochem.(Tokyo), 138, 2005
|
|
2DY7
 
 | |
2DY8
 
 | |
1TBP
 
 | |
2DVW
 
 | |
2DZN
 
 | |
