8CJ5
 
 | |
8CJ8
 
 | |
8OJQ
 
 | |
8OJ9
 
 | |
8OJE
 
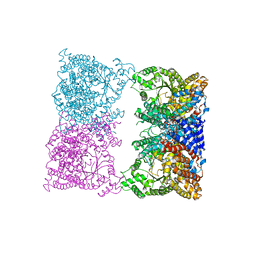 | |
8OJF
 
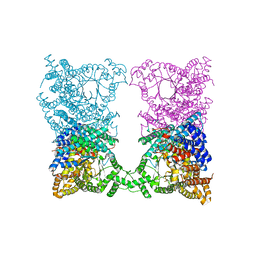 | |
4ELY
 
 | | CCDBVFI:GYRA14EC | | Descriptor: | CHLORIDE ION, CcdB, DNA gyrase subunit A, ... | | Authors: | De Jonge, N, Simic, R, Buts, L, Haesaerts, S, Roelants, K, Garcia-Pino, A, Sterckx, Y, De Greve, H, Lah, J, Loris, R. | | Deposit date: | 2012-04-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Alternative interactions define gyrase specificity in the CcdB family.
Mol.Microbiol., 84, 2012
|
|
4ELZ
 
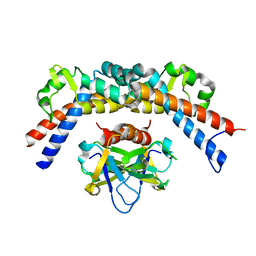 | | CCDBVFI:GYRA14VFI | | Descriptor: | CcdB, DNA gyrase subunit A, GLYCEROL | | Authors: | De Jonge, N, Simic, M, Buts, L, Haesaerts, S, Roelants, K, Garcia-Pino, A, Sterckx, Y, De Greve, H, Lah, J, Loris, R. | | Deposit date: | 2012-04-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Alternative interactions define gyrase specificity in the CcdB family.
Mol.Microbiol., 84, 2012
|
|
8OJZ
 
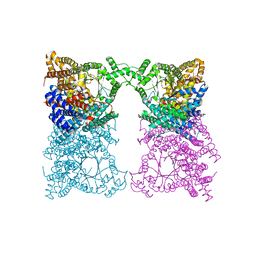 | |
9G2A
 
 | | Staphylococcus aureus MazF in complex with nanobody 4 | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endoribonuclease MazF, ... | | Authors: | Prolic-Kalinsek, M, Zorzini, V, Haesaerts, S, Loris, R. | | Deposit date: | 2024-07-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.046594 Å) | | Cite: | Staphylococcus aureus MazF in complex with nanobody 4.
To Be Published
|
|
9G5Z
 
 | |
9G2G
 
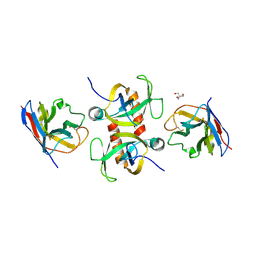 | | Staphylococcus aureus MazF in complex with nanobody 5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoribonuclease MazF, Nanobody 5 | | Authors: | Prolic-Kalinsek, M, Zorzini, V, Haesaerts, S, Loris, R. | | Deposit date: | 2024-07-10 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.80048239 Å) | | Cite: | Nanobody-mediated activation and inhibition of Staphylococcus aureus MazF
To Be Published
|
|
9G4G
 
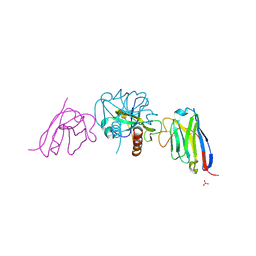 | |
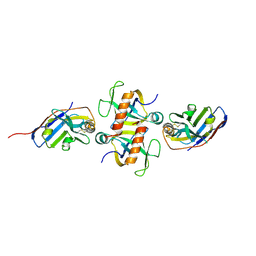
9G1Y
 
 | |
9G48
 
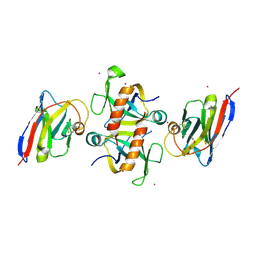 | |
9G5L
 
 | |
9G7K
 
 | |
4MZP
 
 | | MazF from S. aureus crystal form III, C2221, 2.7 A | | Descriptor: | MazF mRNA interferase | | Authors: | Zorzini, V, Loris, R, van Nuland, N.A.J, Cheung, A. | | Deposit date: | 2013-09-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics.
Nucleic Acids Res., 42, 2014
|
|
4MZT
 
 | | MazF from S. aureus crystal form II, C2221, 2.3 A | | Descriptor: | MazF mRNA interferase | | Authors: | Zorzini, V, Loris, R, van Nuland, N.A.J, Cheung, A. | | Deposit date: | 2013-09-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics.
Nucleic Acids Res., 42, 2014
|
|
4MZM
 
 | | MazF from S. aureus crystal form I, P212121, 2.1 A | | Descriptor: | mRNA interferase MazF | | Authors: | Zorzini, V, Loris, R, van Nuland, N.A.J, Cheung, A. | | Deposit date: | 2013-09-30 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics.
Nucleic Acids Res., 42, 2014
|
|
2MF2
 
 | |
5MJE
 
 | | Crystal structure of the HigB2 toxin in complex with Nb8 | | Descriptor: | Cytotoxic translational repressor of toxin-antitoxin stability system, DI(HYDROXYETHYL)ETHER, Nanobody 8, ... | | Authors: | Hadzi, S, Loris, R. | | Deposit date: | 2016-11-30 | | Release date: | 2017-04-05 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Ribosome-dependent Vibrio cholerae mRNAse HigB2 is regulated by a beta-strand sliding mechanism.
Nucleic Acids Res., 45, 2017
|
|
3DD9
 
 | | Structure of DocH66Y dimer | | Descriptor: | Death on curing protein | | Authors: | Garcia-Pino, A, Loris, R. | | Deposit date: | 2008-06-05 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The intrinsically disordered domain of the antitoxin Phd chaperones the toxin Doc against irreversible inactivation and misfolding
J. Biol. Chem., 289, 2014
|
|
5JAA
 
 | |
5JA8
 
 | | Crystal structure of the HigB2 toxin in complex with Nb2 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Hadzi, S, Loris, R. | | Deposit date: | 2016-04-12 | | Release date: | 2017-04-05 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Ribosome-dependent Vibrio cholerae mRNAse HigB2 is regulated by a beta-strand sliding mechanism.
Nucleic Acids Res., 45, 2017
|
|
