2BMK
 
 | |
2GFB
 
 | |
1F3D
 
 | | CATALYTIC ANTIBODY 4B2 IN COMPLEX WITH ITS AMIDINIUM HAPTEN. | | Descriptor: | 2-(4-AMINOBENZYLAMINO)-3,4,5,6-TETRAHYDROPYRIDINIUM, CATALYTIC ANTIBODY 4B2, SULFATE ION | | Authors: | Golinelli-Pimpaneau, B, Goncalves, O, Dintinger, T, Blanchard, D, Knossow, M, Tellier, C. | | Deposit date: | 2000-06-02 | | Release date: | 2000-09-13 | | Last modified: | 2012-07-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural evidence for a programmed general base in the active site of a catalytic antibody.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1WCB
 
 | |
1WC7
 
 | |
6F7T
 
 | |
4AT6
 
 | |
2CMK
 
 | | CYTIDINE MONOPHOSPHATE KINASE IN COMPLEX WITH CYTIDINE-DI-PHOSPHATE | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, PROTEIN (CYTIDINE MONOPHOSPHATE KINASE), SULFATE ION | | Authors: | Golinelli-Pimpaneau, B, Briozzo, P. | | Deposit date: | 1998-09-19 | | Release date: | 1999-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of escherichia coli CMP kinase alone and in complex with CDP: a new fold of the nucleoside monophosphate binding domain and insights into cytosine nucleotide specificity.
Structure, 6, 1998
|
|
3KS0
 
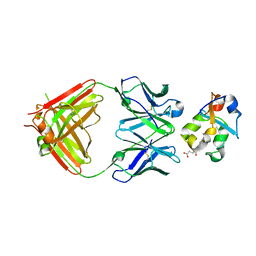 | | Crystal structure of the heme domain of flavocytochrome b2 in complex with Fab B2B4 | | Descriptor: | Cytochrome b2, mitochondrial, Fragment Antigen Binding B2B4, ... | | Authors: | Golinelli-Pimpaneau, B, Lederer, F, Le, K.H.D. | | Deposit date: | 2009-11-20 | | Release date: | 2010-05-26 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence for the functional importance of the heme domain mobility in flavocytochrome b2.
J.Mol.Biol., 400, 2010
|
|
1YED
 
 | |
1YEE
 
 | |
1YEC
 
 | |
4AMV
 
 | |
3OOJ
 
 | |
5MKO
 
 | | [2Fe-2S] cluster containing TtuA in complex with AMP. | | Descriptor: | ADENOSINE MONOPHOSPHATE, FE2/S2 (INORGANIC) CLUSTER, TtuA PH0300, ... | | Authors: | Arragain, S, Bimai, O, Legrand, P, Golinelli-Pimpaneau, B. | | Deposit date: | 2016-12-05 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Nonredox thiolation in tRNA occurring via sulfur activation by a [4Fe-4S] cluster.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MKQ
 
 | | TtuA enzyme containing a [4Fe-4S] | | Descriptor: | IRON/SULFUR CLUSTER, TtuA PH0300, ZINC ION | | Authors: | Arragain, S, Bimai, O, Legrand, P, Golinelli-Pimpaneau, B. | | Deposit date: | 2016-12-05 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Nonredox thiolation in tRNA occurring via sulfur activation by a [4Fe-4S] cluster.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MKP
 
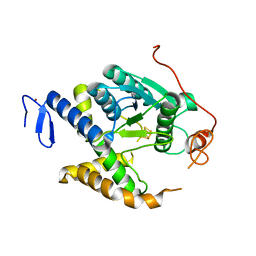 | | Non redox thiolation in transfer RNAs occuring via sulfur activation by a [4Fe-4S] cluster | | Descriptor: | Fe4 H S5, PH0300, ZINC ION | | Authors: | Arragain, S, Bimai, O, Legrand, P, Golinelli-Pimpaneau, B. | | Deposit date: | 2016-12-05 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nonredox thiolation in tRNA occurring via sulfur activation by a [4Fe-4S] cluster.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4WFS
 
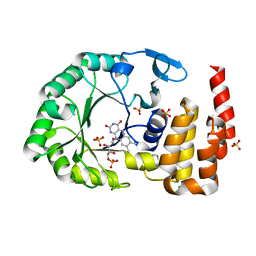 | | Crystal Structure of tRNA-dihydrouridine(20) synthase catalytic domain | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, SULFATE ION, ... | | Authors: | Bou-Nader, C, Pecqueur, L, Kamah, A, Bregeon, D, Golinelli-Pimpaneau, B, Guimaraes, B.G, Fontecave, M, Hamdane, D. | | Deposit date: | 2014-09-17 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | An extended dsRBD is required for post-transcriptional modification in human tRNAs.
Nucleic Acids Res., 43, 2015
|
|
4WFT
 
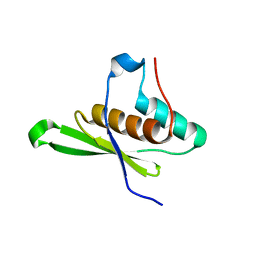 | | Crystal structure of tRNA-dihydrouridine(20) synthase dsRBD domain | | Descriptor: | tRNA-dihydrouridine(20) synthase [NAD(P)+]-like | | Authors: | Bou-Nader, C, Pecqueur, L, Kamah, A, Bregeon, D, Golinelli-Pimpaneau, B, Guimaraes, B.G, Fontecave, M, Hamdane, D. | | Deposit date: | 2014-09-17 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An extended dsRBD is required for post-transcriptional modification in human tRNAs.
Nucleic Acids Res., 43, 2015
|
|
6SCY
 
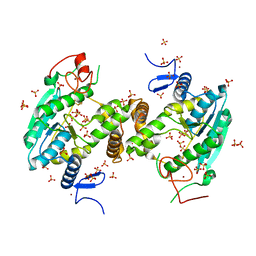 | | U34-tRNA thiolase NcsA from Methanococcus maripaludis with its [4Fe-4S] cluster | | Descriptor: | IRON/SULFUR CLUSTER, SULFATE ION, ZINC ION, ... | | Authors: | Bimai, O, Legrand, P, Golinelli-Pimpaneau, B. | | Deposit date: | 2019-07-25 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The thiolation of uridine 34 in tRNA, which controls protein translation, depends on a [4Fe-4S] cluster in the archaeum Methanococcus maripaludis.
Sci Rep, 13, 2023
|
|
4MPM
 
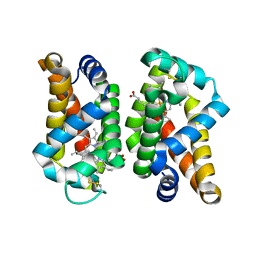 | | Wild-type human neuroglobin | | Descriptor: | Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Guimaraes, B.G, Golinelli-Pimpaneau, B. | | Deposit date: | 2013-09-13 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The crystal structure of wild-type human brain neuroglobin reveals flexibility of the disulfide bond that regulates oxygen affinity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4K22
 
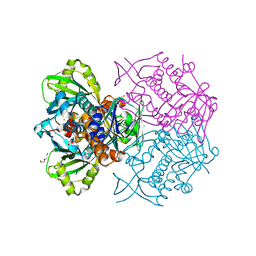 | | Structure of the C-terminal truncated form of E.Coli C5-hydroxylase UBII involved in ubiquinone (Q8) biosynthesis | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Pecqueur, L, Lombard, M, Golinelli-pimpaneau, B, Fontecave, M. | | Deposit date: | 2013-04-07 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ubiI, a New Gene in Escherichia coli Coenzyme Q Biosynthesis, Is Involved in Aerobic C5-hydroxylation.
J.Biol.Chem., 288, 2013
|
|
2PWY
 
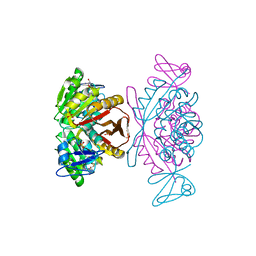 | | Crystal Structure of a m1A58 tRNA methyltransferase | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, tRNA (adenine-N(1)-)-methyltransferase | | Authors: | Barraud, P, Golinelli-Pimpaneau, B, Tisne, C. | | Deposit date: | 2007-05-14 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Thermus thermophilus tRNA m(1)A(58) Methyltransferase and Biophysical Characterization of Its Interaction with tRNA.
J.Mol.Biol., 377, 2008
|
|
2J6H
 
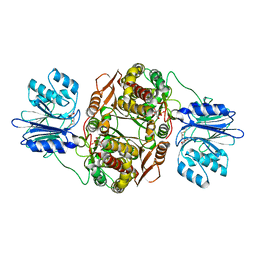 | |
8CP3
 
 | | Apo-LarE in complex with AMP-PNP | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, NAD_synthase domain-containing protein, ... | | Authors: | Zecchin, P, Pecqueur, L, Golinelli-Pimpaneau, B. | | Deposit date: | 2023-03-01 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-based insights into the mechanism of [4Fe-4S]-dependent sulfur insertase LarE.
Protein Sci., 33, 2024
|
|
