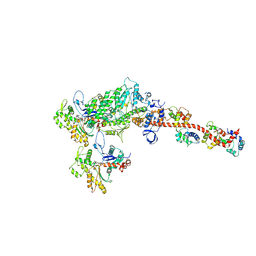
5H53
 
 | |
3J0R
 
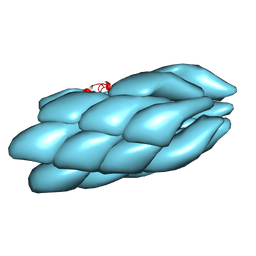 | | Model of a type III secretion system needle based on a 7 Angstrom resolution cryoEM map | | Descriptor: | Protein mxiH | | Authors: | Fujii, T, Cheung, M, Blanco, A, Kato, T, Blocker, A.J, Namba, K. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structure of a type III secretion needle at 7-A resolution provides insights into its assembly and signaling mechanisms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3MFP
 
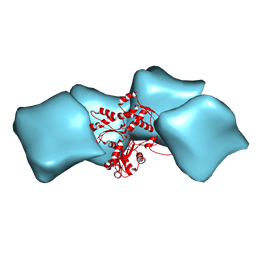 | | Atomic model of F-actin based on a 6.6 angstrom resolution cryoEM map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle | | Authors: | Fujii, T, Iwane, A.H, Yanagida, T, Namba, K. | | Deposit date: | 2010-04-03 | | Release date: | 2010-09-29 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Direct visualization of secondary structures of F-actin by electron cryomicroscopy
Nature, 467, 2010
|
|
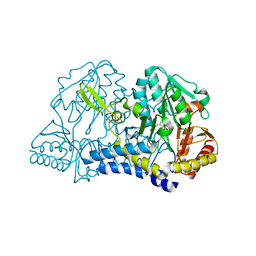
5WRH
 
 | |
1C0N
 
 | | CSDB PROTEIN, NIFS HOMOLOGUE | | Descriptor: | ACETIC ACID, PROTEIN (CSDB PROTEIN), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Fujii, T, Maeda, M, Mihara, H, Kurihara, T, Esaki, N, Hata, Y. | | Deposit date: | 1999-07-17 | | Release date: | 2000-07-17 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a NifS homologue: X-ray structure analysis of CsdB, an Escherichia coli counterpart of mammalian selenocysteine lyase
Biochemistry, 39, 2000
|
|
1XER
 
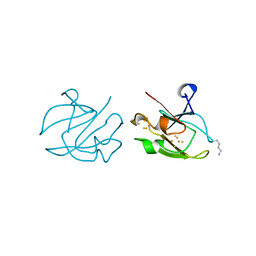 | | STRUCTURE OF FERREDOXIN | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN, ZINC ION | | Authors: | Fujii, T, Hata, Y, Moriyama, H, Wakagi, T, Tanaka, N, Oshima, T. | | Deposit date: | 1996-08-28 | | Release date: | 1997-09-04 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel zinc-binding centre in thermoacidophilic archaeal ferredoxins.
Nat.Struct.Biol., 3, 1996
|
|
3W5S
 
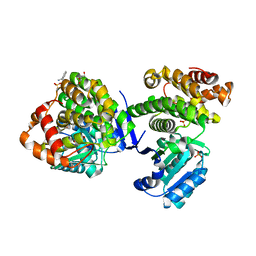 | | Crystal Structure of Maleylacetate Reductase from Rhizobium sp. strain MTP-10005 | | Descriptor: | BENZAMIDINE, GLYCEROL, Maleylacetate reductase, ... | | Authors: | Fujii, T, Ogawa, A, Goda, Y, Yamauchi, T, Yoshida, M, Oikawa, T, Hata, Y. | | Deposit date: | 2013-02-06 | | Release date: | 2014-02-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Structure of Maleylacetate Reductase from Rhizobium sp. strain MTP-10005
To be Published
|
|
1J3U
 
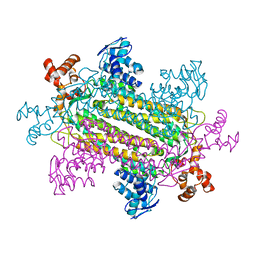 | |
3A69
 
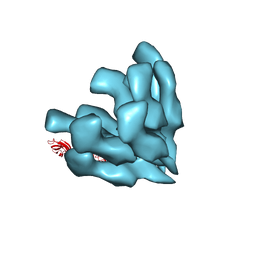 | |
1UHA
 
 | | Crystal Structure of Pokeweed Lectin-D2 | | Descriptor: | CALCIUM ION, lectin-D2 | | Authors: | Fujii, T, Hayashida, M, Hamasu, M, Ishiguro, M, Hata, Y. | | Deposit date: | 2003-06-27 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of two lectins from the roots of pokeweed (Phytolacca americana).
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1ULN
 
 | | Crystal Structure of Pokeweed Lectin-D1 | | Descriptor: | lectin-D | | Authors: | Fujii, T, Hayashida, M, Hamasu, M, Ishiguro, M, Hata, Y. | | Deposit date: | 2003-09-16 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of two lectins from the roots of pokeweed (Phytolacca americana).
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6KN8
 
 | | Structure of human cardiac thin filament in the calcium bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Fujii, T, Yamada, Y, Namba, K. | | Deposit date: | 2019-08-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cardiac muscle thin filament structures reveal calcium regulatory mechanism.
Nat Commun, 11, 2020
|
|
6KN7
 
 | | Structure of human cardiac thin filament in the calcium free state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Fujii, T, Yamada, Y, Namba, K. | | Deposit date: | 2019-08-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cardiac muscle thin filament structures reveal calcium regulatory mechanism.
Nat Commun, 11, 2020
|
|
1QH9
 
 | | ENZYME-PRODUCT COMPLEX OF L-2-HALOACID DEHALOGENASE | | Descriptor: | 2-HALOACID DEHALOGENASE, LACTIC ACID | | Authors: | Li, Y.-F, Hata, Y, Fujii, T, Kurihara, T, Esaki, N. | | Deposit date: | 1999-05-12 | | Release date: | 2000-05-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of L-2-Haloacid Dehalogenase Complexed with a Reaction Product Reveals the Mechanism of Intermediate Hydrolysis in Dehalogenase
To be Published
|
|
1I29
 
 | | CRYSTAL STRUCTURE OF CSDB COMPLEXED WITH L-PROPARGYLGLYCINE | | Descriptor: | (2S)-2-aminobut-3-ynoic acid, CSDB, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mihara, H, Fujii, T, Kurihara, T, Hata, Y, Esaki, N. | | Deposit date: | 2001-02-07 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of external aldimine of Escherichia coli CsdB, an IscS/NifS homolog: implications for its specificity toward
selenocysteine.
J.BIOCHEM.(TOKYO), 131, 2002
|
|
1JUD
 
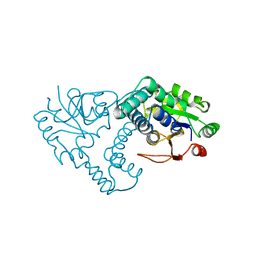 | | L-2-HALOACID DEHALOGENASE | | Descriptor: | L-2-HALOACID DEHALOGENASE | | Authors: | Hisano, T, Hata, Y, Fujii, T, Liu, J.-Q, Kurihara, T, Esaki, N, Soda, K. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of L-2-haloacid dehalogenase from Pseudomonas sp. YL. An alpha/beta hydrolase structure that is different from the alpha/beta hydrolase fold.
J.Biol.Chem., 271, 1996
|
|
3J6P
 
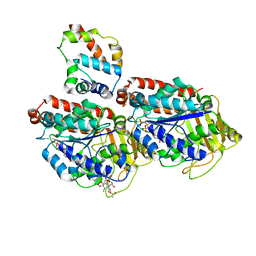 | | Pseudo-atomic model of dynein microtubule binding domain-tubulin complex based on a cryoEM map | | Descriptor: | Dynein heavy chain, cytoplasmic, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Uchimura, S, Fujii, T, Takazaki, H, Ayukawa, R, Nishikawa, Y, Minoura, I, Hachikubo, Y, Kurisu, G, Sutoh, K, Kon, T, Namba, K, Muto, E. | | Deposit date: | 2014-03-20 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | A flipped ion pair at the dynein-microtubule interface is critical for dynein motility and ATPase activation
J.Cell Biol., 208, 2015
|
|
2Z36
 
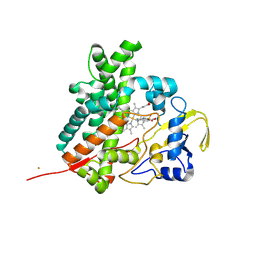 | | Crystal structure of cytochrome P450 MoxA from Nonomuraea recticatena (CYP105) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cytochrome P450 type compactin 3'',4''-hydroxylase, FE (III) ION, ... | | Authors: | Yasutake, Y, Fujii, Y, Fujii, T, Arisawa, A, Tamura, T. | | Deposit date: | 2007-06-02 | | Release date: | 2007-08-21 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of cytochrome P450 MoxA from Nonomuraea recticatena (CYP105)
Biochem.Biophys.Res.Commun., 361, 2007
|
|
9L3J
 
 | | Crystal structure of endo-processive xyloglucanase Xeg5A from Aspergillus oryzae with GXG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoside hydrolase superfamily, ... | | Authors: | Nakamichi, Y, Shimada, N, Watanabe, M, Fujii, T, Matsuzawa, T. | | Deposit date: | 2024-12-18 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural insights into substrate recognition of tri-modular xyloglucanase from Aspergillus oryzae.
J.Struct.Biol., 217, 2025
|
|
9L3P
 
 | | Crystal structure of endo-processive xyloglucanase Xeg5A E285A from Aspergillus oryzae with GLLX/L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoside hydrolase superfamily, alpha-D-mannopyranose, ... | | Authors: | Nakamichi, Y, Shimada, N, Watanabe, M, Fujii, T, Matsuzawa, T. | | Deposit date: | 2024-12-19 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural insights into substrate recognition of tri-modular xyloglucanase from Aspergillus oryzae.
J.Struct.Biol., 217, 2025
|
|
9L3D
 
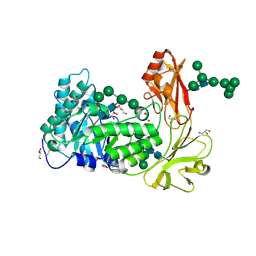 | | Crystal structure of endo-processive xyloglucanase Xeg5A from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Nakamichi, Y, Shimada, N, Watanabe, M, Fujii, T, Matsuzawa, T. | | Deposit date: | 2024-12-18 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into substrate recognition of tri-modular xyloglucanase from Aspergillus oryzae.
J.Struct.Biol., 217, 2025
|
|
9L3O
 
 | | Crystal structure of endo-processive xyloglucanase Xeg5A from Aspergillus oryzae with XXLG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Glycoside hydrolase superfamily, ... | | Authors: | Nakamichi, Y, Shimada, N, Watanabe, M, Fujii, T, Matsuzawa, T. | | Deposit date: | 2024-12-19 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into substrate recognition of tri-modular xyloglucanase from Aspergillus oryzae.
J.Struct.Biol., 217, 2025
|
|
3J82
 
 | | Electron cryo-microscopy of DNGR-1 in complex with F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Hanc, P, Fujii, T, Yamada, Y, Huotari, J, Schulz, O, Ahrens, S, Kjaer, S, Way, M, Namba, K, Reis e Sousa, C. | | Deposit date: | 2014-09-25 | | Release date: | 2015-05-20 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structure of the Complex of F-Actin and DNGR-1, a C-Type Lectin Receptor Involved in Dendritic Cell Cross-Presentation of Dead Cell-Associated Antigens.
Immunity, 42, 2015
|
|
5XDH
 
 | | His/DOPA ligated cytochrome c from an anammox organism KSU-1 | | Descriptor: | ACETATE ION, HEME C, Putative cytochrome c, ... | | Authors: | Hira, D, Kitamura, R, Nakamura, T, Yamagata, Y, Furukawa, K, Fujii, T. | | Deposit date: | 2017-03-28 | | Release date: | 2018-03-28 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Anammox Organism KSU-1 Expresses a Novel His/DOPA Ligated Cytochrome c.
J. Mol. Biol., 430, 2018
|
|
1ZRM
 
 | | CRYSTAL STRUCTURE OF THE REACTION INTERMEDIATE OF L-2-HALOACID DEHALOGENASE WITH 2-CHLORO-N-BUTYRATE | | Descriptor: | L-2-HALOACID DEHALOGENASE, butanoic acid | | Authors: | Li, Y.-F, Hata, Y, Fujii, T, Hisano, T, Nishihara, M, Kurihara, T, Esaki, N. | | Deposit date: | 1998-03-03 | | Release date: | 1999-03-23 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of reaction intermediates of L-2-haloacid dehalogenase and implications for the reaction mechanism.
J.Biol.Chem., 273, 1998
|
|
