4X2V
 
 | |
4X2W
 
 | |
6G6S
 
 | | Crystal structure of human Acinus RNA recognition motif domain | | Descriptor: | Apoptotic chromatin condensation inducer in the nucleus | | Authors: | Fernandes, H, Czapinska, H, Grudziaz, K, Bujnicki, J.M, Nowacka, M. | | Deposit date: | 2018-04-03 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of human Acinus RNA recognition motif domain.
PeerJ, 6, 2018
|
|
4X2X
 
 | |
4X2Y
 
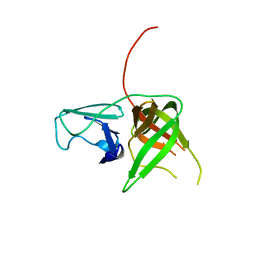 | | Crystal structure of a chimeric Murine Norovirus NS6 protease (inactive C139A mutant) in which the P4-P4 prime residues of the cleavage junction in the extended C-terminus have been replaced by the corresponding residues from the NS2-3 junction. | | Descriptor: | NS6 Protease,NS6 Protease | | Authors: | Leen, E.N, Pfeil, M.-P, Fernandes, H, Curry, S. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.417 Å) | | Cite: | Structure determination of Murine Norovirus NS6 proteases with C-terminal extensions designed to probe protease-substrate interactions.
Peerj, 3, 2015
|
|
3IE5
 
 | | Crystal structure of Hyp-1 protein from Hypericum perforatum (St John's wort) involved in hypericin biosynthesis | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Michalska, K, Fernandes, H, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.688 Å) | | Cite: | Crystal structure of Hyp-1, a St. John's wort protein implicated in the biosynthesis of hypericin
J.Struct.Biol., 169, 2010
|
|
6IA2
 
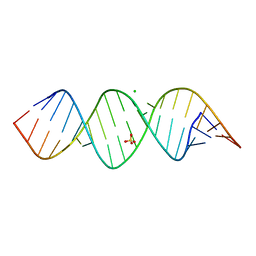 | | Crystal structure of a self-complementary RNA duplex recognized by Com | | Descriptor: | CHLORIDE ION, RNA (5'-R(*AP*GP*AP*GP*AP*AP*CP*CP*CP*GP*GP*AP*GP*UP*UP*CP*CP*CP*U)-3'), SULFATE ION | | Authors: | Nowacka, M, Fernandes, H, Kiliszek, A, Bernat, A, Lach, G, Bujnicki, J.M. | | Deposit date: | 2018-11-26 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Specific interaction of zinc finger protein Com with RNA and the crystal structure of a self-complementary RNA duplex recognized by Com.
Plos One, 14, 2019
|
|
3TYI
 
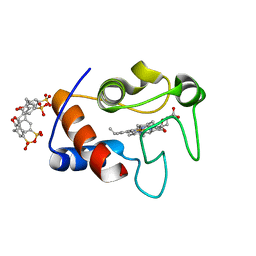 | | Crystal Structure of Cytochrome c - p-Sulfonatocalix[4]arene Complexes | | Descriptor: | 25,26,27,28-tetrahydroxypentacyclo[19.3.1.1~3,7~.1~9,13~.1~15,19~]octacosa-1(25),3(28),4,6,9(27),10,12,15(26),16,18,21,23-dodecaene-5,11,17,23-tetrasulfonic acid, Cytochrome c iso-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mc Govern, R.E, Fernandes, H, Khan, A.R, Crowley, P.B. | | Deposit date: | 2011-09-26 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Protein camouflage in cytochrome c-calixarene complexes.
Nat Chem, 4, 2012
|
|
3E85
 
 | | Crystal Structure of Pathogenesis-related Protein LlPR-10.2B from yellow lupine in complex with Diphenylurea | | Descriptor: | 1,3-DIPHENYLUREA, PR10.2B, SODIUM ION | | Authors: | Fernandes, H.C, Bujacz, G, Bujacz, A, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2008-08-19 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cytokinin-induced structural adaptability of a Lupinus luteus PR-10 protein.
Febs J., 276, 2009
|
|
2QIM
 
 | | Crystal Structure of Pathogenesis-related Protein LlPR-10.2B from yellow lupine in complex with Cytokinin | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, CALCIUM ION, GLYCEROL, ... | | Authors: | Fernandes, H.C, Pasternak, O, Bujacz, G, Bujacz, A, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2007-07-05 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Lupinus luteus pathogenesis-related protein as a reservoir for cytokinin.
J.Mol.Biol., 378, 2008
|
|
4GY9
 
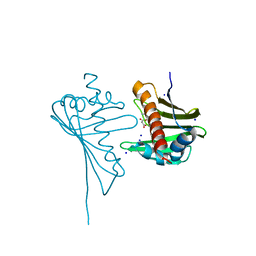 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with N6-isopentenyladenine (2iP) | | Descriptor: | MALONATE ION, MtN13 protein, N-(3-METHYLBUT-2-EN-1-YL)-9H-PURIN-6-AMINE, ... | | Authors: | Ruszkowski, M, Sikorski, M, Jaskolski, M. | | Deposit date: | 2012-09-05 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The landscape of cytokinin binding by a plant nodulin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5I8F
 
 | | Crystal structure of St. John's wort Hyp-1 protein in complex with melatonin | | Descriptor: | GLYCEROL, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Phenolic oxidative coupling protein, ... | | Authors: | Sliwiak, J, Dauter, Z, Jaskolski, M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Hyp-1, a Hypericum perforatum PR-10 Protein, in Complex with Melatonin.
Front Plant Sci, 7, 2016
|
|
5MXB
 
 | | Crystal structure of yellow lupin LLPR-10.2B protein in complex with melatonin | | Descriptor: | Class 10 plant pathogenesis-related protein, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, SODIUM ION, ... | | Authors: | Sliwiak, J, Sikorski, M, Jaskolski, M. | | Deposit date: | 2017-01-22 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | PR-10 proteins as potential mediators of melatonin-cytokinin cross-talk in plants: crystallographic studies of LlPR-10.2B isoform from yellow lupine.
FEBS J., 285, 2018
|
|
5MXW
 
 | | Crystal structure of yellow lupin LLPR-10.2B protein in complex with melatonin and trans-zeatin. | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, Class 10 plant pathogenesis-related protein, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, ... | | Authors: | Sliwiak, J, Sikorski, M, Jaskolski, M. | | Deposit date: | 2017-01-25 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | PR-10 proteins as potential mediators of melatonin-cytokinin cross-talk in plants: crystallographic studies of LlPR-10.2B isoform from yellow lupine.
FEBS J., 285, 2018
|
|
4JHG
 
 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with trans-zeatin | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, MALONATE ION, MtN13 protein, ... | | Authors: | Ruszkowski, M, Tusnio, K, Ciesielska, A, Brzezinski, K, Dauter, M, Dauter, Z, Sikorski, M, Jaskolski, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The landscape of cytokinin binding by a plant nodulin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4JHI
 
 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with N6-benzyladenine | | Descriptor: | MtN13 protein, N-BENZYL-9H-PURIN-6-AMINE, SODIUM ION | | Authors: | Ruszkowski, M, Sikorski, M, Jaskolski, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The landscape of cytokinin binding by a plant nodulin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4JHH
 
 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with kinetin | | Descriptor: | MALONATE ION, MtN13 protein, N-(FURAN-2-YLMETHYL)-7H-PURIN-6-AMINE, ... | | Authors: | Ruszkowski, M, Sikorski, M, Jaskolski, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The landscape of cytokinin binding by a plant nodulin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4PSB
 
 | |
4Q0K
 
 | | Crystal Structure of Phytohormone Binding Protein from Medicago truncatula in complex with gibberellic acid (GA3) | | Descriptor: | GIBBERELLIN A3, GLYCEROL, PHYTOHORMONE BINDING PROTEIN MTPHBP | | Authors: | Ciesielska, A, Barciszewski, J, Ruszkowski, M, Jaskolski, M, Sikorski, M. | | Deposit date: | 2014-04-02 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Specific binding of gibberellic acid by Cytokinin-Specific Binding Proteins: a new aspect of plant hormone-binding proteins with the PR-10 fold.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
