7PKW
 
 | | Crystal structure of VIRB8-like OrfG central and C-terminal domains of Streptococcus thermophilus ICESt3 (Gram positive conjugative type IV secretion system). | | Descriptor: | GLYCEROL, Putative transfer protein, SULFATE ION | | Authors: | Favier, F, Didierjean, C, Cappele, J, Douzi, B, Leblond-Bourget, N. | | Deposit date: | 2021-08-27 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.835 Å) | | Cite: | Crystal structure of VIRB8-like OrfG central and C-terminal domains of Streptococcus thermophilus ICESt3 (Gram positive conjugative type IV secretion system).
To Be Published
|
|
4USS
 
 | | Populus trichocarpa glutathione transferase X1-1 (GHR1), complexed with glutathione | | Descriptor: | GLUTATHIONE, GLUTATHIONYL HYDROQUINONE REDUCTASE, PHOSPHATE ION | | Authors: | Lallement, P.A, Meux, E, Gualberto, J.M, Dumaracay, S, Favier, F, Didierjean, C, Saul, F, Haouz, A, Morel-Rouhier, M, Gelhaye, E, Rouhier, N, Hecker, A. | | Deposit date: | 2014-07-13 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glutathionyl-Hydroquinone Reductases from Poplar are Plastidial Proteins that Deglutathionylate Both Reduced and Oxidized Glutathionylated Quinones.
FEBS Lett., 589, 2015
|
|
1DLA
 
 | | NOVEL NADPH-BINDING DOMAIN REVEALED BY THE CRYSTAL STRUCTURE OF ALDOSE REDUCTASE | | Descriptor: | ALDOSE REDUCTASE | | Authors: | Rondeau, J.-M, Tete-Favier, F, Podjarny, A, Reymann, J.-M, Barth, P, Biellmann, J.-F, Moras, D. | | Deposit date: | 1993-02-08 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel NADPH-binding domain revealed by the crystal structure of aldose reductase.
Nature, 355, 1992
|
|
6ZGN
 
 | | Crystal structure of VirB8-like OrfG central domain of Streptococcus thermophilus ICESt3; a putative assembly factor of a gram positive conjugative Type IV secretion system. | | Descriptor: | Putative transfer protein | | Authors: | Cappele, J, Mohamad-Ali, A, Leblond-Bourget, N, Payot-Lacroix, S, Mathiot, S, Didierjean, C, Favier, F, Douzi, B. | | Deposit date: | 2020-06-19 | | Release date: | 2021-04-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biochemical Analysis of OrfG: The VirB8-like Component of the Conjugative Type IV Secretion System of ICE St3 From Streptococcus thermophilus .
Front Mol Biosci, 8, 2021
|
|
1FF3
 
 | | STRUCTURE OF THE PEPTIDE METHIONINE SULFOXIDE REDUCTASE FROM ESCHERICHIA COLI | | Descriptor: | PEPTIDE METHIONINE SULFOXIDE REDUCTASE, SULFATE ION | | Authors: | Tete-Favier, F, Cobessi, D, Boschi-Muller, S, Azza, S, Branlant, G, Aubry, A. | | Deposit date: | 2000-07-25 | | Release date: | 2000-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Escherichia coli peptide methionine sulphoxide reductase at 1.9 A resolution.
Structure Fold.Des., 8, 2000
|
|
3HCG
 
 | |
8PFE
 
 | |
3PPU
 
 | |
7PKA
 
 | | Synechocystis sp. PCC6803 glutathione transferase Chi 1, GSOH bound | | Descriptor: | Glutathione S-transferase, POTASSIUM ION, S-Hydroxy-Glutathione | | Authors: | Didierjean, C, Mocchetti, E, Hecker, A, Favier, F. | | Deposit date: | 2021-08-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure and functions of glutathione transferase Chi 1 from cyanobacterium Synechocystis sp. PCC 6803
To Be Published
|
|
5LKD
 
 | | Crystal structure of the Xi glutathione transferase ECM4 from Saccharomyces cerevisiae in complex with glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase omega-like 2 | | Authors: | Schwartz, M, Didierjean, C, Hecker, A, Girardet, J.M, Morel-Rouhier, M, Gelhaye, E, Favier, F. | | Deposit date: | 2016-07-22 | | Release date: | 2016-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Saccharomyces cerevisiae ECM4, a Xi-Class Glutathione Transferase that Reacts with Glutathionyl-(hydro)quinones.
Plos One, 11, 2016
|
|
3HCH
 
 | | Structure of the C-terminal domain (MsrB) of Neisseria meningitidis PilB (complex with substrate) | | Descriptor: | (2S)-2-(acetylamino)-N-methyl-4-[(R)-methylsulfinyl]butanamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRIC ACID, ... | | Authors: | Ranaivoson, F.M, Kauffmann, B, Favier, F. | | Deposit date: | 2009-05-06 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methionine sulfoxide reductase B displays a high level of flexibility.
J.Mol.Biol., 394, 2009
|
|
3HCJ
 
 | |
3HCI
 
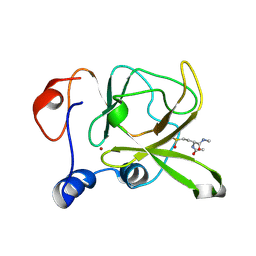 | | Structure of MsrB from Xanthomonas campestris (complex-like form) | | Descriptor: | (2S)-2-(acetylamino)-N-methyl-4-[(R)-methylsulfinyl]butanamide, CALCIUM ION, Peptide methionine sulfoxide reductase, ... | | Authors: | Ranaivoson, F.M, Kauffmann, B, Favier, F. | | Deposit date: | 2009-05-06 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Methionine sulfoxide reductase B displays a high level of flexibility.
J.Mol.Biol., 394, 2009
|
|
5LKB
 
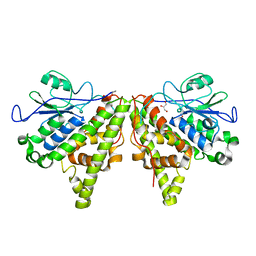 | | Crystal structure of the Xi glutathione transferase ECM4 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Glutathione S-transferase omega-like 2 | | Authors: | Schwartz, M, Didierjean, C, Hecker, A, Girardet, J.M, Morel-Rouhier, M, Gelhaye, E, Favier, F. | | Deposit date: | 2016-07-22 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Saccharomyces cerevisiae ECM4, a Xi-Class Glutathione Transferase that Reacts with Glutathionyl-(hydro)quinones.
Plos One, 11, 2016
|
|
1EUH
 
 | | APO FORM OF A NADP DEPENDENT ALDEHYDE DEHYDROGENASE FROM STREPTOCOCCUS MUTANS | | Descriptor: | NADP DEPENDENT NON PHOSPHORYLATING GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, SULFATE ION | | Authors: | Cobessi, D, Tete-Favier, F, Marchal, S, Branlant, G, Aubry, A. | | Deposit date: | 1998-11-05 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Apo and holo crystal structures of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans.
J.Mol.Biol., 290, 1999
|
|
3S1I
 
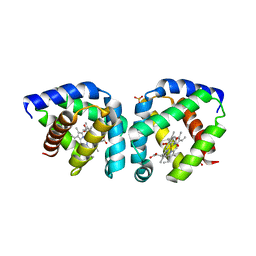 | | Crystal structure of oxygen-bound hell's gate globin I | | Descriptor: | Hemoglobin-like flavoprotein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
3S1J
 
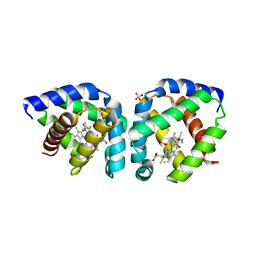 | | Crystal structure of acetate-bound hell's gate globin I | | Descriptor: | ACETATE ION, Hemoglobin-like flavoprotein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
6HPE
 
 | | Crystal structure of glutathione transferase Omega 3S from Trametes versicolor in complex with the glutathione adduct of phenethyl-isothiocyanate | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2018-09-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of Trametes versicolor glutathione transferase Omega 3S bound to its conjugation product glutathionyl-phenethylthiocarbamate reveals plasticity of its active site.
Protein Sci., 28, 2019
|
|
6HTA
 
 | | Crystal structure of glutathione transferase Xi 3 mutant C56S from Trametes versicolor | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Glutathione transferase Xi 3 C56S | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2018-10-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Trametes versicolor glutathione transferase Xi 3, a dual Cys-GST with catalytic specificities of both Xi and Omega classes.
FEBS Lett., 592, 2018
|
|
4LMW
 
 | |
4LMV
 
 | |
2FY6
 
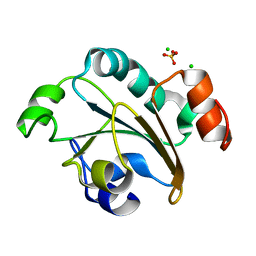 | | Structure of the N-terminal domain of Neisseria meningitidis PilB | | Descriptor: | CHLORIDE ION, Peptide methionine sulfoxide reductase msrA/msrB, SULFATE ION | | Authors: | Ranaivoson, F.M, Kauffmann, B, Neiers, F, Boschi-Muller, S, Branlant, G, Favier, F. | | Deposit date: | 2006-02-07 | | Release date: | 2006-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray Structure of the N-terminal Domain of PILB from Neisseria meningitidis Reveals a Thioredoxin-fold
J.Mol.Biol., 358, 2006
|
|
3BQG
 
 | |
3BQE
 
 | |
3BQH
 
 | |
