5O6Y
 
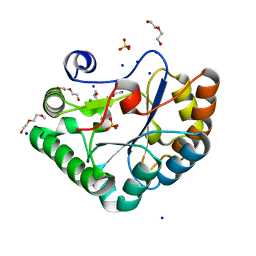 | | Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase in complex with 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide, ... | | Authors: | Fadouloglou, V.E, Kotsifaki, D, Kokkinidis, M. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase in complex with 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide
To Be Published
|
|
1O9Y
 
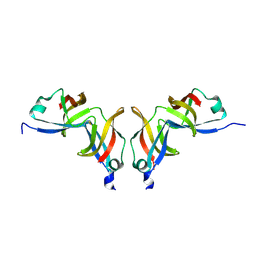 | |
2IXD
 
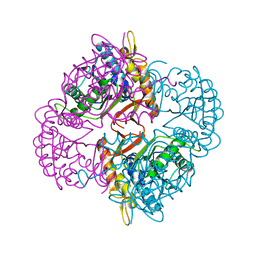 | | Crystal structure of the putative deacetylase BC1534 from Bacillus cereus | | Descriptor: | ACETATE ION, LMBE-RELATED PROTEIN, ZINC ION | | Authors: | Fadouloglou, V.E, Bouriotis, V, Kokkinidis, M. | | Deposit date: | 2006-07-07 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Bczbp, a Zinc-Binding Protein from Bacillus Cereus
FEBS J., 274, 2007
|
|
4HD5
 
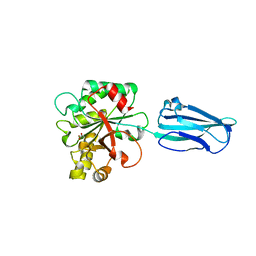 | | Crystal Structure of BC0361, a polysaccharide deacetylase from Bacillus cereus | | Descriptor: | ACETATE ION, Polysaccharide deacetylase, ZINC ION | | Authors: | Fadouloglou, V.E, Kokkinidis, M, Glykos, N.M. | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure determination through homology modelling and torsion-angle simulated annealing: application to a polysaccharide deacetylase from Bacillus cereus.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4L1G
 
 | |
6G2U
 
 | | Crystal structure of the human glutamate dehydrogenase 2 (hGDH2) | | Descriptor: | CHLORIDE ION, Glutamate dehydrogenase 2, mitochondrial, ... | | Authors: | Fadouloglou, V.F, Dimovasili, C, Providaki, M, Kotsifaki, D, Sarrou, I, Plaitakis, A, Zaganas, I, Kokkinidis, M. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.934287 Å) | | Cite: | Crystal structure of glutamate dehydrogenase 2, a positively selected novel human enzyme involved in brain biology and cancer pathophysiology.
J.Neurochem., 2021
|
|
