1GWD
 
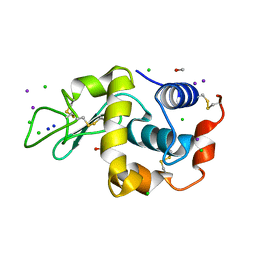 | | Tri-iodide derivative of hen egg-white lysozyme | | Descriptor: | 1,2-ETHANEDIOL, CARBON MONOXIDE, CHLORIDE ION, ... | | Authors: | Evans, G, Bricogne, G. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Triiodide Derivatization and Combinatorial Counter-Ion Replacement: Two Methods for Enhancing Phasing Signal Using Laboratory Cu Kalpha X-Ray Equipment
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GW9
 
 | |
1GWG
 
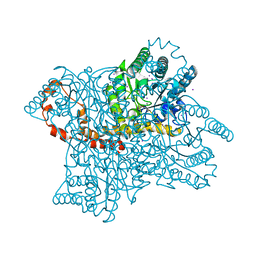
 | | Tri-iodide derivative of apoferritin | | Descriptor: | CADMIUM ION, FERRITIN LIGHT CHAIN, IODIDE ION | | Authors: | Evans, G, Bricogne, G. | | Deposit date: | 2002-03-15 | | Release date: | 2002-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Triiodide Derivatization and Combinatorial Counter-Ion Replacement: Two Methods for Enhancing Phasing Signal Using Laboratory Cu Kalpha X-Ray Equipment
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GWA
 
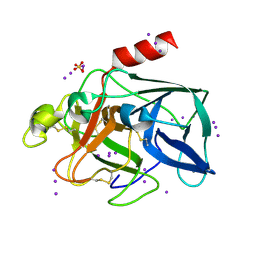 | | Triiodide derivative of porcine pancreas elastase | | Descriptor: | CALCIUM ION, ELASTASE 1, IODIDE ION, ... | | Authors: | Evans, G, Bricogne, G. | | Deposit date: | 2002-03-13 | | Release date: | 2002-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Triiodide Derivatization and Combinatorial Counter-Ion Replacement: Two Methods for Enhancing Phasing Signal Using Laboratory Cu Kalpha X-Ray Equipment
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6ZET
 
 | | Crystal structure of proteinase K nanocrystals by electron diffraction with a 20 micrometre C2 condenser aperture | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Evans, G, Zhang, P, Beale, E.V, Waterman, D.G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.701 Å) | | Cite: | A Workflow for Protein Structure Determination From Thin Crystal Lamella by Micro-Electron Diffraction.
Front Mol Biosci, 7, 2020
|
|
6ZEV
 
 | | Crystal structure of proteinase K lamellae by electron diffraction with a 20 micrometre C2 condenser aperture | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Evans, G, Zhang, P, Beale, E.V, Waterman, D.G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.4 Å) | | Cite: | A Workflow for Protein Structure Determination From Thin Crystal Lamella by Micro-Electron Diffraction.
Front Mol Biosci, 7, 2020
|
|
6ZEU
 
 | | Crystal structure of proteinase K lamella by electron diffraction with a 50 micrometre C2 condenser aperture | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Evans, G, Zhang, P, Beale, E.V, Waterman, D.G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.004 Å) | | Cite: | A Workflow for Protein Structure Determination From Thin Crystal Lamella by Micro-Electron Diffraction.
Front Mol Biosci, 7, 2020
|
|
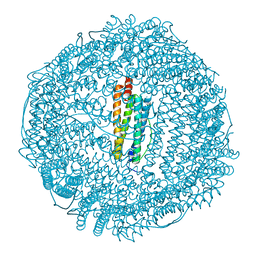
8QPH
 
 | | Crystal structure of Lymantria dispar CPV14 polyhedra 14 crystals | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | Authors: | Trincao, J, Warren, A, Crawshaw, A, Sutton, G, Stuart, D, Evans, G. | | Deposit date: | 2023-10-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | VMXm - A sub-micron focus macromolecular crystallography beamline at Diamond Light Source.
J.Synchrotron Radiat., 31, 2024
|
|
8QQC
 
 | | Crystal structure of Lymantria dispar CPV14 polyhedra single crystal | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | Authors: | Trincao, J, Warren, A, Crawshaw, A, Sutton, G, Stuart, D, Evans, G. | | Deposit date: | 2023-10-04 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | VMXm - A sub-micron focus macromolecular crystallography beamline at Diamond Light Source.
J.Synchrotron Radiat., 31, 2024
|
|
2WUX
 
 | | the crystal structure of recombinant baculovirus polyhedra | | Descriptor: | POLYHEDRIN | | Authors: | Ji, X, Sutton, G, Evans, G, Axford, D, Owen, R, Stuart, D.I. | | Deposit date: | 2009-10-10 | | Release date: | 2009-12-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | How Baculovirus Polyhedra Fit Square Pegs Into Round Holes to Robustly Package Viruses.
Embo J., 29, 2010
|
|
2WUY
 
 | | the crystal structure of wild-type baculovirus polyhedra | | Descriptor: | POLYHEDRIN | | Authors: | Ji, X, Sutton, G, Evans, G, Axford, D, Owen, R, Stuart, D.I. | | Deposit date: | 2009-10-10 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | How Baculovirus Polyhedra Fit Square Pegs Into Round Holes to Robustly Package Viruses.
Embo J., 29, 2010
|
|
5LNS
 
 | | Crystal structure of Arabidopsis thaliana Pdx1-R5P complex | | Descriptor: | PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, RIBULOSE-5-PHOSPHATE | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
5LNU
 
 | | Crystal structure of Arabidopsis thaliana Pdx1-I320 complex | | Descriptor: | (4~{S})-4-azanyl-5-oxidanyl-pent-1-en-3-one, PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
5LNT
 
 | | Crystal structure of Arabidopsis thaliana Pdx1K166R-preI320 complex | | Descriptor: | PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.1, [(~{E},4~{S})-4-azanyl-3-oxidanylidene-pent-1-enyl] dihydrogen phosphate | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
5LNR
 
 | | Crystal structure of Arabidopsis thaliana Pdx1-PLP complex | | Descriptor: | GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal 5'-phosphate synthase subunit PDX1.3 | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
5LNV
 
 | | Crystal structure of Arabidopsis thaliana Pdx1-I320 complex from multiple crystals | | Descriptor: | (4~{S})-4-azanyl-5-oxidanyl-pent-1-en-3-one, PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
5LNW
 
 | | Crystal structure of Arabidopsis thaliana Pdx1-I320-G3P complex | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, GLYCEROL, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.00001 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5KAI
 
 | | NH3-bound RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 2.8 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.80000925 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
4YCR
 
 | | Structure determination of an integral membrane protein at room temperature from crystals in situ | | Descriptor: | Tellurite resistance protein TehA homolog, octyl beta-D-glucopyranoside | | Authors: | Axford, D, Hu, N.J, Foadi, J, Choudhury, H.G, Iwata, S, Beis, K, Evans, G, Alguel, Y. | | Deposit date: | 2015-02-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure determination of an integral membrane protein at room temperature from crystals in situ.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4ZQX
 
 | | A revised partiality model and post-refinement algorithm for X-ray free-electron laser data | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | Authors: | Ginn, H.M, Brewster, A.S, Hattne, J, Evans, G, Wagner, A, Grimes, J, Sauter, N.K, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-05-11 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A revised partiality model and post-refinement algorithm for X-ray free-electron laser data.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5A9B
 
 | | Crystal structure of Bombyx mori CPV1 polyhedra base domain deleted mutant | | Descriptor: | POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A9C
 
 | | Crystal structure of Antheraea mylitta CPV4 polyhedra base domain deleted mutant | | Descriptor: | POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A98
 
 | | Crystal structure of Trichoplusia ni CPV15 polyhedra | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A96
 
 | | Crystal structure of Lymantria dispar CPV14 polyhedra | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
