2JK1
 
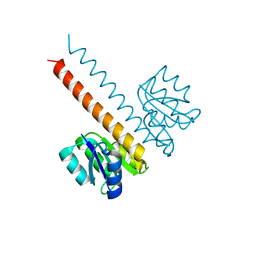 | | Crystal structure of the wild-type HupR receiver domain | | Descriptor: | HYDROGENASE TRANSCRIPTIONAL REGULATORY PROTEIN HUPR1, MAGNESIUM ION | | Authors: | Davies, K.M, Lowe, E.D, Venien-Bryan, C, Johnson, L.N. | | Deposit date: | 2008-05-26 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Hupr Receiver Domain Crystal Structure in its Nonphospho and Inhibitory Phospho States.
J.Mol.Biol., 385, 2009
|
|
2VUI
 
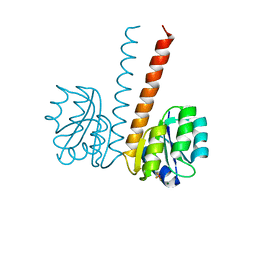 | | Crystal structure of the HupR receiver domain in inhibitory phospho- state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, HYDROGENASE TRANSCRIPTIONAL REGULATORY PROTEIN HUPR1, MAGNESIUM ION | | Authors: | Davies, K.M, Lowe, E.D, Venien-Bryan, C, Johnson, L.N. | | Deposit date: | 2008-05-26 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Hupr Receiver Domain Crystal Structure in its Nonphospho and Inhibitory Phospho States.
J.Mol.Biol., 385, 2009
|
|
2VUH
 
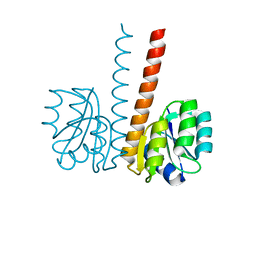 | |
4B2Q
 
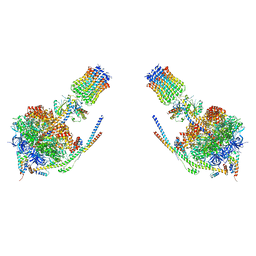 | |
6EDH
 
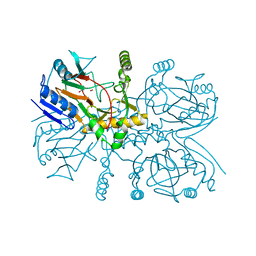 | | Taurine:2OG dioxygenase (TauD) bound to the vanadyl ion, taurine, and succinate | | Descriptor: | 2-AMINOETHANESULFONIC ACID, ACETATE ION, Alpha-ketoglutarate-dependent taurine dioxygenase, ... | | Authors: | Davis, K.M, Altmyer, M, Boal, A.K. | | Deposit date: | 2018-08-09 | | Release date: | 2019-08-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.73000407 Å) | | Cite: | Structure of a Ferryl Mimic in the Archetypal Iron(II)- and 2-(Oxo)-glutarate-Dependent Dioxygenase, TauD.
Biochemistry, 58, 2019
|
|
5V1S
 
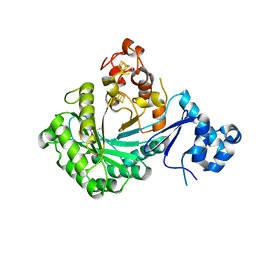 | |
5V1T
 
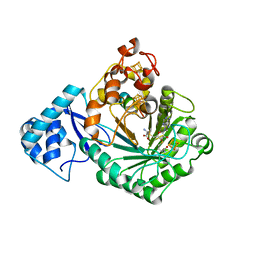 | | Crystal structure of Streptococcus suis SuiB bound to precursor peptide SuiA | | Descriptor: | IRON/SULFUR CLUSTER, METHIONINE, Radical SAM, ... | | Authors: | Davis, K.M, Bacik, J.P, Ando, N. | | Deposit date: | 2017-03-02 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the peptide-modifying radical SAM enzyme SuiB elucidate the basis of substrate recognition.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5V1Q
 
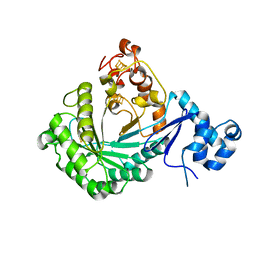 | | Crystal structure of Streptococcus suis SuiB | | Descriptor: | IRON/SULFUR CLUSTER, Radical SAM | | Authors: | Davis, K.M, Bacik, J.P, Ando, N. | | Deposit date: | 2017-03-02 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the peptide-modifying radical SAM enzyme SuiB elucidate the basis of substrate recognition.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7LKC
 
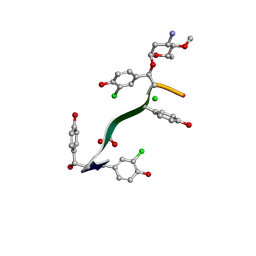 | | Crystal Structure of Keratinimicin A | | Descriptor: | (2~{S},4~{S},5~{R},6~{S})-4-azanyl-5-methoxy-6-methyl-oxan-2-ol, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Davis, K.M, Jeffrey, P.D, Seyedsayamdost, M.R. | | Deposit date: | 2021-02-02 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Potent and specific antibiotic combination therapy against Clostridioides difficile.
Nat.Chem.Biol., 20, 2024
|
|
7LTB
 
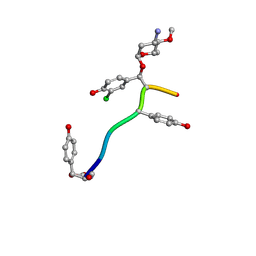 | | Crystal Structure of Keratinicyclin B | | Descriptor: | (2~{S},4~{S},5~{R},6~{S})-4-azanyl-5-methoxy-6-methyl-oxan-2-ol, 3-ammonio-2,3,6-trideoxy-alpha-L-arabino-hexopyranose-(1-2)-beta-D-glucopyranose, FORMIC ACID, ... | | Authors: | Davis, K.M, Jeffrey, P.D, Seyedsayamdost, M.R. | | Deposit date: | 2021-02-19 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Potent and specific antibiotic combination therapy against Clostridioides difficile.
Nat.Chem.Biol., 20, 2024
|
|
6VP5
 
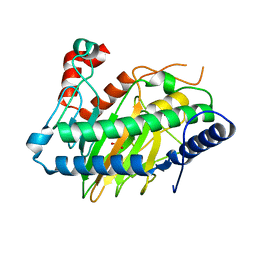 | | Ethylene forming enzyme (EFE) D191E variant in complex with Fe(II), L-arginine, and 2OG | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ... | | Authors: | Davis, K.M, Copeland, R.A, Boal, A.K. | | Deposit date: | 2020-02-01 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | An Iron(IV)-Oxo Intermediate Initiating l-Arginine Oxidation but Not Ethylene Production by the 2-Oxoglutarate-Dependent Oxygenase, Ethylene-Forming Enzyme.
J.Am.Chem.Soc., 143, 2021
|
|
6VP4
 
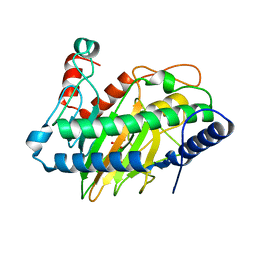 | | Ethylene forming enzyme (EFE) in complex with Fe(II), L-arginine, and 2OG | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ... | | Authors: | Davis, K.M, Copeland, R.A, Boal, A.K. | | Deposit date: | 2020-02-01 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | An Iron(IV)-Oxo Intermediate Initiating l-Arginine Oxidation but Not Ethylene Production by the 2-Oxoglutarate-Dependent Oxygenase, Ethylene-Forming Enzyme.
J.Am.Chem.Soc., 143, 2021
|
|
6OWG
 
 | | Structure of a synthetic beta-carboxysome shell, T=4 | | Descriptor: | Ethanolamine utilization protein EutN/carboxysome structural protein Ccml, Microcompartments protein | | Authors: | Sutter, M, Laughlin, T.G, Davies, K.M, Kerfeld, C.A. | | Deposit date: | 2019-05-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of a Syntheticbeta-Carboxysome Shell.
Plant Physiol., 181, 2019
|
|
6OWF
 
 | | Structure of a synthetic beta-carboxysome shell, T=3 | | Descriptor: | Ethanolamine utilization protein EutN/carboxysome structural protein Ccml, Microcompartments protein | | Authors: | Sutter, M, Laughlin, T.G, Davies, K.M, Kerfeld, C.A. | | Deposit date: | 2019-05-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a Syntheticbeta-Carboxysome Shell.
Plant Physiol., 181, 2019
|
|
6NBX
 
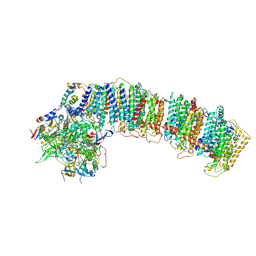 | | T.elongatus NDH (data-set 2) | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase chain 4 1, NAD(P)H-quinone oxidoreductase subunit 1, ... | | Authors: | Laughlin, T.G, Bayne, A, Trempe, J.-F, Savage, D.F, Davies, K.M. | | Deposit date: | 2018-12-10 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Nature, 566, 2019
|
|
6NBY
 
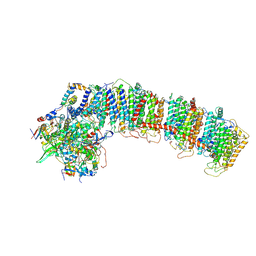 | | T.elongatus NDH (composite model) | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase chain 4 1, NAD(P)H-quinone oxidoreductase subunit 1, ... | | Authors: | Laughlin, T.G, Bayne, A, Trempe, J.-F, Savage, D.F, Davies, K.M. | | Deposit date: | 2018-12-10 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Nature, 566, 2019
|
|
6NBQ
 
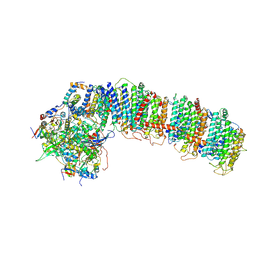 | | T.elongatus NDH (data-set 1) | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase chain 4 1, NAD(P)H-quinone oxidoreductase subunit 2, ... | | Authors: | Laughlin, T.G, Bayne, A, Trempe, J.-F, Savage, D.F, Davies, K.M. | | Deposit date: | 2018-12-09 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Nature, 566, 2019
|
|
8FBX
 
 | | Selenosugar synthase SenB from Variovorax paradoxus | | Descriptor: | CHLORIDE ION, SODIUM ION, Selenosugar synthase SenB | | Authors: | Ireland, K.A, Davis, K.M. | | Deposit date: | 2022-11-30 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Characterization and Ligand-Induced Conformational Changes of SenB, a Se-Glycosyltransferase Involved in Selenoneine Biosynthesis.
Biochemistry, 62, 2023
|
|
8G6C
 
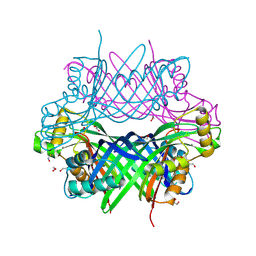 | | GTP Cyclohydrolase-IB with manganese | | Descriptor: | 1,2-ETHANEDIOL, GTP cyclohydrolase FolE2, MANGANESE (II) ION | | Authors: | McWhorter, K.L, Amaya Lopez, C.Y, Davis, K.M. | | Deposit date: | 2023-02-14 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Combatting melioidosis with chemical synthetic lethality.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8G8V
 
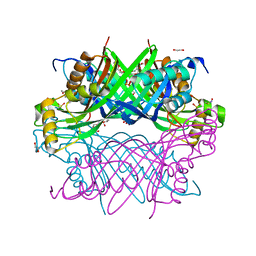 | | GTP Cyclohydrolase-IB with sodium | | Descriptor: | 1,2-ETHANEDIOL, GTP cyclohydrolase FolE2, SODIUM ION, ... | | Authors: | McWhorter, K.L, Jhunjhunwala, R, Davis, K.M. | | Deposit date: | 2023-02-20 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Combatting melioidosis with chemical synthetic lethality.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8F91
 
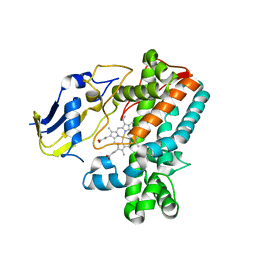 | |
8U41
 
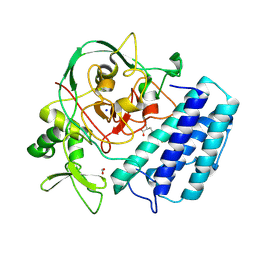 | |
8U42
 
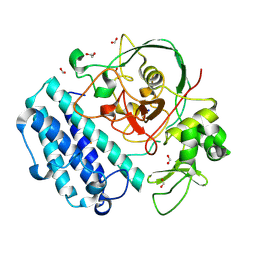 | | OvsA from Halomonas utahensis, a selenoxide synthase involved in ovoselenol biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, FORMIC ACID, ... | | Authors: | Ireland, K.A, Davis, K.M. | | Deposit date: | 2023-09-08 | | Release date: | 2024-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of the selenium-containing antioxidant ovoselenol derived from convergent evolution.
Nat.Chem., 16, 2024
|
|
8UX5
 
 | |
8VIH
 
 | | EgtB-IV from Crocosphaera subtropica, an ergothioneine-biosynthetic type IV sulfoxide synthase in complex with hercynine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Ireland, K.A, Davis, K.M. | | Deposit date: | 2024-01-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the convergent evolution of sulfoxide synthase EgtB-IV, an ergothioneine-biosynthetic homolog of ovothiol synthase OvoA.
Structure, 32, 2024
|
|
