6HQ2
 
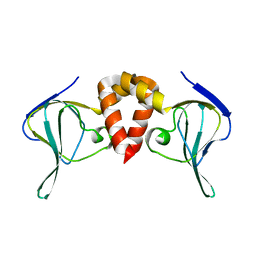 | | Structure of EAL Enzyme Bd1971 - apo form | | Descriptor: | EAL Enzyme Bd1971 | | Authors: | Lovering, A.L, Cadby, I.T. | | Deposit date: | 2018-09-24 | | Release date: | 2019-07-31 | | Last modified: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Nucleotide signaling pathway convergence in a cAMP-sensing bacterial c-di-GMP phosphodiesterase.
Embo J., 38, 2019
|
|
6HQ5
 
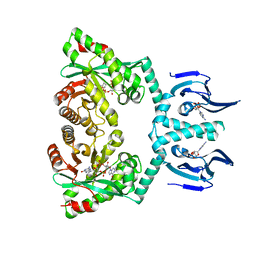 | | Structure of EAL Enzyme Bd1971 - cAMP and cyclic-di-GMP bound form | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CALCIUM ION, ... | | Authors: | Lovering, A.L, Cadby, I.T. | | Deposit date: | 2018-09-24 | | Release date: | 2019-07-31 | | Last modified: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Nucleotide signaling pathway convergence in a cAMP-sensing bacterial c-di-GMP phosphodiesterase.
Embo J., 38, 2019
|
|
6HQ7
 
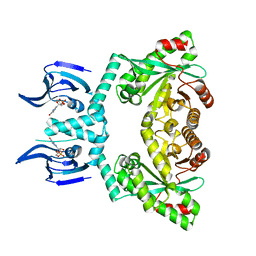 | | Structure of EAL Enzyme Bd1971 - cGMP bound form | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, EAL Enzyme Bd1971, MAGNESIUM ION | | Authors: | Lovering, A.L, Cadby, I.T. | | Deposit date: | 2018-09-24 | | Release date: | 2019-07-31 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Nucleotide signaling pathway convergence in a cAMP-sensing bacterial c-di-GMP phosphodiesterase.
Embo J., 38, 2019
|
|
6HQ4
 
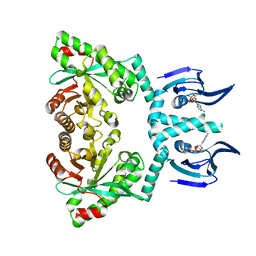 | | Structure of EAL enzyme Bd1971 - cAMP bound form | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, EAL Enzyme Bd1971, MAGNESIUM ION | | Authors: | Lovering, A.L, Cadby, I.T. | | Deposit date: | 2018-09-24 | | Release date: | 2019-07-31 | | Last modified: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Nucleotide signaling pathway convergence in a cAMP-sensing bacterial c-di-GMP phosphodiesterase.
Embo J., 38, 2019
|
|
6HQ3
 
 | |
6SBW
 
 | | CdbA Form One | | Descriptor: | CdbA | | Authors: | Lovering, A.L, Cadby, I.T. | | Deposit date: | 2019-07-22 | | Release date: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | CdbA is a DNA-binding protein and c-di-GMP receptor important for nucleoid organization and segregation in Myxococcus xanthus.
Nat Commun, 11, 2020
|
|
6SBX
 
 | | CdbA Form Two | | Descriptor: | CdbA | | Authors: | Lovering, A.L, Cadby, I.T. | | Deposit date: | 2019-07-22 | | Release date: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | CdbA is a DNA-binding protein and c-di-GMP receptor important for nucleoid organization and segregation in Myxococcus xanthus.
Nat Commun, 11, 2020
|
|
6SAR
 
 | | E coli BepA/YfgC | | Descriptor: | Beta-barrel assembly-enhancing protease, SULFATE ION, ZINC ION | | Authors: | Lovering, A.L, Cadby, I.T. | | Deposit date: | 2019-07-17 | | Release date: | 2020-08-26 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-Function Characterization of the Conserved Regulatory Mechanism of the Escherichia coli M48 Metalloprotease BepA.
J.Bacteriol., 203, 2020
|
|
5CEA
 
 | |
5CED
 
 | | Penicillin G Acylated Bd3459 Predatory Endopeptidase from Bdellovibrio bacteriovorus in complex with immunity protein Bd3460 | | Descriptor: | Bd3459, Bd3460, OPEN FORM - PENICILLIN G | | Authors: | Lovering, A.L, Cadby, I.T, Lambert, C, Sockett, R.E. | | Deposit date: | 2015-07-06 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Ankyrin-mediated self-protection during cell invasion by the bacterial predator Bdellovibrio bacteriovorus.
Nat Commun, 6, 2015
|
|
5CER
 
 | | Bd0816 Predatory Endopeptidase from Bdellovibrio bacteriovorus in complex with immunity protein Bd3460 | | Descriptor: | 1,2-ETHANEDIOL, Bd0816, Bd3460 | | Authors: | Lovering, A.L, Cadby, I.T, Lambert, C, Sockett, R.E. | | Deposit date: | 2015-07-07 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Ankyrin-mediated self-protection during cell invasion by the bacterial predator Bdellovibrio bacteriovorus.
Nat Commun, 6, 2015
|
|
5CEC
 
 | | Bd3459 Predatory Endopeptidase from Bdellovibrio bacteriovorus in complex with immunity protein Bd3460 | | Descriptor: | Bd3459, Bd3460, SULFATE ION | | Authors: | Lovering, A.L, Cadby, I.T, Lambert, C, Sockett, R.E. | | Deposit date: | 2015-07-06 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Ankyrin-mediated self-protection during cell invasion by the bacterial predator Bdellovibrio bacteriovorus.
Nat Commun, 6, 2015
|
|
5CEB
 
 | | Bd3459 Predatory Endopeptidase from Bdellovibrio bacteriovorus, K38M form | | Descriptor: | Bd3459 | | Authors: | Lovering, A.L, Cadby, I.T, Lambert, C, Sockett, R.E. | | Deposit date: | 2015-07-06 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Ankyrin-mediated self-protection during cell invasion by the bacterial predator Bdellovibrio bacteriovorus.
Nat Commun, 6, 2015
|
|
6HBZ
 
 | | Bdellovibrio bacteriovorus DgcB Full-length | | Descriptor: | 1,2-ETHANEDIOL, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CHLORIDE ION, ... | | Authors: | Lovering, A.L, Meek, R.W. | | Deposit date: | 2018-08-13 | | Release date: | 2019-08-28 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural basis for activation of a diguanylate cyclase required for bacterial predation in Bdellovibrio.
Nat Commun, 10, 2019
|
|
6HC0
 
 | |
6SCG
 
 | | Structure of AdhE form 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Aldehyde-alcohol dehydrogenase, ... | | Authors: | Lovering, A.L, Bragginton, E. | | Deposit date: | 2019-07-24 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-resolution structure of the alcohol dehydrogenase domain of the bifunctional bacterial enzyme AdhE.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6SCI
 
 | | Structure of AdhE form 1 | | Descriptor: | Aldehyde-alcohol dehydrogenase, FE (III) ION | | Authors: | Lovering, A.L, Bragginton, E. | | Deposit date: | 2019-07-24 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution structure of the alcohol dehydrogenase domain of the bifunctional bacterial enzyme AdhE.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7NTG
 
 | | Bdellovibrio bacteriovorus PGI in complex with fructose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, FRUCTOSE -6-PHOSPHATE, Glucose-6-phosphate isomerase | | Authors: | Meek, R.W, Lovering, A.L. | | Deposit date: | 2021-03-09 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Bdellovibrio bacteriovorus phosphoglucose isomerase structures reveal novel rigidity in the active site of a selected subset of enzymes upon substrate binding.
Open Biology, 11, 2021
|
|
7O0A
 
 | | Bdellovibrio bacteriovorus PGI in P1211 spacegroup | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glucose-6-phosphate isomerase | | Authors: | Meek, R.W, Lovering, A.L. | | Deposit date: | 2021-03-26 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Bdellovibrio bacteriovorus phosphoglucose isomerase structures reveal novel rigidity in the active site of a selected subset of enzymes upon substrate binding.
Open Biology, 11, 2021
|
|
7NSS
 
 | | Bdellovibrio bacteriovorus PGI in P3121 spacegroup | | Descriptor: | 1,2-ETHANEDIOL, Glucose-6-phosphate isomerase | | Authors: | Meek, R.W, Lovering, A.L. | | Deposit date: | 2021-03-08 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Bdellovibrio bacteriovorus phosphoglucose isomerase structures reveal novel rigidity in the active site of a selected subset of enzymes upon substrate binding.
Open Biology, 11, 2021
|
|
7O21
 
 | |
6GFV
 
 | | M tuberculosis LpqI | | Descriptor: | Probable conserved lipoprotein LpqI | | Authors: | Moynihan, P.J, Lovering, A.L. | | Deposit date: | 2018-05-02 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The hydrolase LpqI primes mycobacterial peptidoglycan recycling.
Nat Commun, 10, 2019
|
|
6GKI
 
 | | Structure of E coli MlaC in Variously Loaded States | | Descriptor: | BROMIDE ION, GLYCEROL, Probable phospholipid-binding protein MlaC | | Authors: | Knowles, T.J, Lovering, A.L. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system.
Nat Microbiol, 4, 2019
|
|
6HC1
 
 | |
6ZNR
 
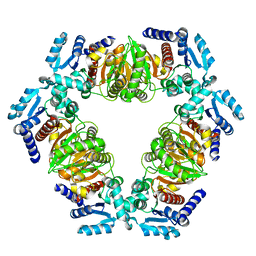 | | MaeB PTA domain R535A mutant | | Descriptor: | Malate dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.217 Å) | | Cite: | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
