1KX6
 
 | |
1MRT
 
 | | CONFORMATION OF CD-7 METALLOTHIONEIN-2 FROM RAT LIVER IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2 | | Authors: | Braun, W, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformation of [Cd7]-metallothionein-2 from rat liver in aqueous solution determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 203, 1988
|
|
1MRB
 
 | | THREE-DIMENSIONAL STRUCTURE OF RABBIT LIVER CD7 METALLOTHIONEIN-2A IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2A | | Authors: | Braun, W, Arseniev, A, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of rabbit liver [Cd7]metallothionein-2a in aqueous solution determined by nuclear magnetic resonance.
J.Mol.Biol., 201, 1988
|
|
2MRB
 
 | | THREE-DIMENSIONAL STRUCTURE OF RABBIT LIVER CD-7 METALLOTHIONEIN-2A IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2A | | Authors: | Braun, W, Arseniev, A, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of rabbit liver [Cd7]metallothionein-2a in aqueous solution determined by nuclear magnetic resonance.
J.Mol.Biol., 201, 1988
|
|
2MRT
 
 | | CONFORMATION OF CD-7 METALLOTHIONEIN-2 FROM RAT LIVER IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2 | | Authors: | Braun, W, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformation of [Cd7]-metallothionein-2 from rat liver in aqueous solution determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 203, 1988
|
|
1MHU
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF HUMAN [113CD7] METALLOTHIONEIN-2 IN SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2 | | Authors: | Braun, W, Messerle, B.A, Schaeffer, A, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of human [113Cd7]metallothionein-2 in solution determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 214, 1990
|
|
2MHU
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF HUMAN [113CD7] METALLOTHIONEIN-2 IN SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2 | | Authors: | Braun, W, Messerle, B.A, Schaeffer, A, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of human [113Cd7]metallothionein-2 in solution determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 214, 1990
|
|
1CXR
 
 | |
3CYS
 
 | | DETERMINATION OF THE NMR SOLUTION STRUCTURE OF THE CYCLOPHILIN A-CYCLOSPORIN A COMPLEX | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Spitzfaden, C, Braun, W, Wider, G, Widmer, H, Wuthrich, K. | | Deposit date: | 1994-02-28 | | Release date: | 1994-08-31 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Determination of the NMR Solution Structure of the Cyclophilin A-Cyclosporin a Complex.
J.Biomol.NMR, 4, 1994
|
|
3AIT
 
 | |
4AIT
 
 | |
2AIT
 
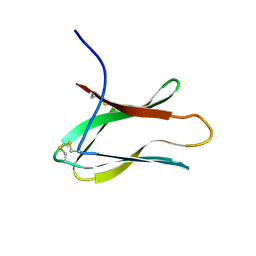 | | DETERMINATION OF THE COMPLETE THREE-DIMENSIONAL STRUCTURE OF THE ALPHA-AMYLASE INHIBITOR TENDAMISTAT IN AQUEOUS SOLUTION BY NUCLEAR MAGNETIC RESONANCE AND DISTANCE GEOMETRY | | Descriptor: | TENDAMISTAT | | Authors: | Kline, A.D, Braun, W, Guntert, P, Billeter, M, Wuthrich, K. | | Deposit date: | 1989-05-24 | | Release date: | 1990-04-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Determination of the complete three-dimensional structure of the alpha-amylase inhibitor tendamistat in aqueous solution by nuclear magnetic resonance and distance geometry.
J.Mol.Biol., 204, 1988
|
|
1T50
 
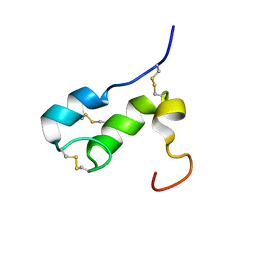 | | NMR SOLUTION STRUCTURE OF APLYSIA ATTRACTIN | | Descriptor: | Attractin | | Authors: | Ravindranath, G, Xu, Y, Schein, C.H, Rajaratnam, K, Painter, S.D, Nagle, G.T, Braun, W. | | Deposit date: | 2004-04-30 | | Release date: | 2004-05-11 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Attractin, a Water-Borne Pheromone from the Mollusk Aplysia Attractin
Biochemistry, 42, 2003
|
|
1NH5
 
 | | AUTOMATIC ASSIGNMENT OF NMR DATA AND DETERMINATION OF THE PROTEIN STRUCTURE OF A NEW WORLD SCORPION NEUROTOXIN USING NOAH/DIAMOD | | Descriptor: | Neurotoxin 5 | | Authors: | Xu, Y, Jablonsky, M.J, Jackson, P.L, Krishna, N.R, Braun, W. | | Deposit date: | 2002-12-18 | | Release date: | 2003-01-07 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Automatic assignment of NOESY Cross peaks and determination of the protein structure of a new world scorpion neurotoxin Using NOAH/DIAMOD
J.Magn.Reson., 148, 2001
|
|
4MT2
 
 | |
2BBL
 
 | |
2BBP
 
 | |
1PIT
 
 | |
