4WMH
 
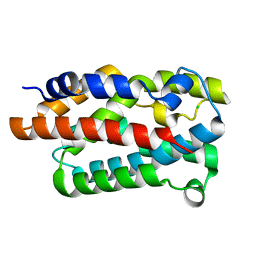 | |
3IA8
 
 | |
3EMM
 
 | | X-ray structure of protein from Arabidopsis thaliana AT1G79260 with Bound Heme | | Descriptor: | 1,2-ETHANEDIOL, PROTOPORPHYRIN IX CONTAINING FE, Uncharacterized protein At1g79260 | | Authors: | Bianchetti, C.M, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-09-24 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.358 Å) | | Cite: | The structure and NO binding properties of the nitrophorin-like heme-binding protein from Arabidopsis thaliana gene locus At1g79260.1.
Proteins, 78, 2010
|
|
3NDZ
 
 | |
3NDY
 
 | |
3QDE
 
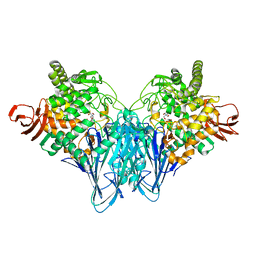 | | The structure of Cellobiose phosphorylase from Clostridium thermocellum in complex with phosphate | | Descriptor: | Cellobiose phosphorylase, PHOSPHATE ION, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM | | Authors: | Bianchetti, C.M, Elsen, N.L, Horn, M.K, Fox, B.G, Phillips Jr, G.N. | | Deposit date: | 2011-01-18 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of cellobiose phosphorylase from Clostridium thermocellum in complex with phosphate.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4TYV
 
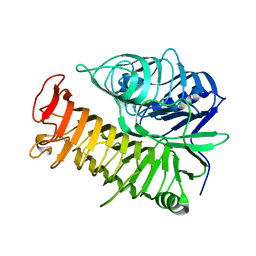 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4TZ3
 
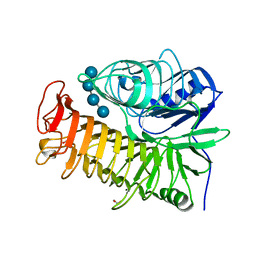 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritetraose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose, ... | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4TZ1
 
 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritriose | | Descriptor: | Putative secreted protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4TZ5
 
 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
2Q32
 
 | | Crystal structure of human heme oxygenase-2 C127A (HO-2) | | Descriptor: | Heme oxygenase 2, OXTOXYNOL-10 | | Authors: | Bianchetti, C.M, Bingman, C.A, Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparison of Apo- and Heme-bound Crystal Structures of a Truncated Human Heme Oxygenase-2.
J.Biol.Chem., 282, 2007
|
|
2QPP
 
 | | Crystal structure of human heme oxygenase-2 C127A (HO-2) with bound heme | | Descriptor: | Heme oxygenase 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bianchetti, C.M, Bingman, C.A, Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-07-24 | | Release date: | 2007-08-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Comparison of Apo- and Heme-bound Crystal Structures of a Truncated Human Heme Oxygenase-2.
J.Biol.Chem., 282, 2007
|
|
2RGZ
 
 | | Ensemble refinement of the protein crystal structure of human heme oxygenase-2 C127A (HO-2) with bound heme | | Descriptor: | Heme oxygenase 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bianchetti, C.M, Bingman, C.A, Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-10-05 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Comparison of Apo- and Heme-bound Crystal Structures of a Truncated Human Heme Oxygenase-2.
J.Biol.Chem., 282, 2007
|
|
4ILT
 
 | | Structure of the dioxygenase domain of SACTE_2871, a novel dioxygenase carbohydrate-binding protein fusion from the cellulolytic bacterium Streptomyces sp. SirexAA-E | | Descriptor: | CHLORIDE ION, FE (III) ION, Intradiol ring-cleavage dioxygenase | | Authors: | Bianchetti, C.M, Takasuka, T.E, Bergeman, L.F, Harmann, C.H, Fox, B.G. | | Deposit date: | 2012-12-31 | | Release date: | 2013-05-15 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Fusion of Dioxygenase and Lignin-binding Domains in a Novel Secreted Enzyme from Cellulolytic Streptomyces sp. SirexAA-E.
J.Biol.Chem., 288, 2013
|
|
4ILV
 
 | | Structure of the dioxygenase domain of SACTE_2871, a novel dioxygenase carbohydrate-binding protein fusion from the cellulolytic bacterium Streptomyces sp. SirexAA-E | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Intradiol ring-cleavage dioxygenase | | Authors: | Bianchetti, C.M, Takasuka, T.E, Bergeman, L.F, Harmann, C.H, Fox, B.G. | | Deposit date: | 2013-01-01 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Fusion of Dioxygenase and Lignin-binding Domains in a Novel Secreted Enzyme from Cellulolytic Streptomyces sp. SirexAA-E.
J.Biol.Chem., 288, 2013
|
|
4PF0
 
 | | Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-04-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4PEY
 
 | | Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritriose | | Descriptor: | Putative secreted protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-04-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4PEZ
 
 | | Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritetraose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose, ... | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-04-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4PEX
 
 | | Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-04-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4PEW
 
 | | Structure of sacteLam55A from Streptomyces sp. SirexAA-E | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Putative secreted protein | | Authors: | Bianchetti, C.M, Takasuka, T.E, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-04-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
3IAU
 
 | | The structure of the processed form of threonine deaminase isoform 2 from Solanum lycopersicum | | Descriptor: | ACETATE ION, POLYETHYLENE GLYCOL (N=34), SULFATE ION, ... | | Authors: | Bianchetti, C.M, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-07-14 | | Release date: | 2009-07-28 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Adaptive evolution of threonine deaminase in plant defense against insect herbivores.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6Q1I
 
 | |
6PZ7
 
 | | GH5-4 broad specificity endoglucanase from Clostridium acetobutylicum | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Endoglucanase family 5 | | Authors: | Bianchetti, C.M, Bingman, C.A, Fox, B.G. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6UI3
 
 | | GH5-4 broad specificity endoglucanase from Clostridum cellulovorans | | Descriptor: | 1,2-ETHANEDIOL, Cellulase | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6WQV
 
 | | GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose | | Descriptor: | 1,2-ETHANEDIOL, Endoglucanase, NITRATE ION, ... | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
