2WY4
 
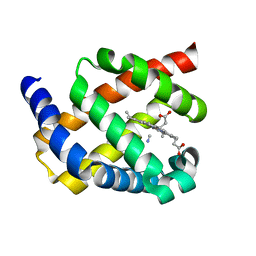 | | Structure of bacterial globin from Campylobacter jejuni at 1.35 A resolution | | Descriptor: | CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SINGLE DOMAIN HAEMOGLOBIN | | Authors: | Barynin, V.V, Sedelnikova, S.E, Shepherd, M, Wu, G, Poole, R.K, Rice, D.W. | | Deposit date: | 2009-11-11 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Single-Domain Globin from the Pathogenic Bacterium Campylobacter Jejuni: Novel D-Helix Conformation, Proximal Hydrogen Bonding that Influences Ligand Binding, and Peroxidase-Like Redox Properties.
J.Biol.Chem., 285, 2010
|
|
4PW9
 
 | |
4PWA
 
 | |
4PW3
 
 | |
6U3K
 
 | | The crystal structure of 4-(pyridin-2-yl)benzoate-bound CYP199A4 | | Descriptor: | 4-(pyridin-2-yl)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical Techniques for Distinguishing Ligand Binding Modes in Cytochrome P450 Monooxygenases.
Biochemistry, 59, 2020
|
|
5WA0
 
 | |
7N14
 
 | | Crystal structure of 4-(1H-1,2,4-triazol-1-yl)benzoic acid-bound CYP199A4 | | Descriptor: | 4-(1H-1,2,4-triazol-1-yl)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2021-05-26 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.537 Å) | | Cite: | To Be, or Not to Be, an Inhibitor: A Comparison of Azole Interactions with and Oxidation by a Cytochrome P450 Enzyme.
Inorg.Chem., 61, 2022
|
|
7KCS
 
 | | The crystal structure of 4-vinylbenzoate-bound wild-type CYP199A4 | | Descriptor: | 4-ethenylbenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Coleman, T, Bruning, J.B, Bell, S.G. | | Deposit date: | 2020-10-07 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Understanding the Mechanistic Requirements for Efficient and Stereoselective Alkene Epoxidation by a Cytochrome P450 Enzyme
Acs Catalysis, 11, 2021
|
|
7L5S
 
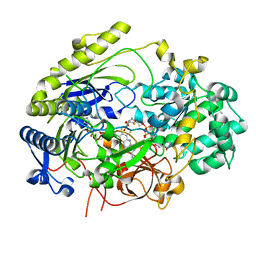 | | Crystal Structure of Haemophilus influenzae MtsZ at pH 5.5 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, MOLYBDENUM ATOM, OXYGEN ATOM, ... | | Authors: | Struwe, M.A, Luo, Z, Kappler, U, Kobe, B. | | Deposit date: | 2020-12-22 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Active site architecture reveals coordination sphere flexibility and specificity determinants in a group of closely related molybdoenzymes.
J.Biol.Chem., 296, 2021
|
|
7L5I
 
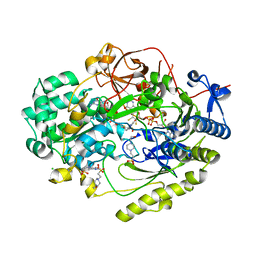 | | Crystal Structure of Haemophilus influenzae MtsZ at pH 7.0 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Struwe, M.A, Luo, Z, Kappler, U, Kobe, B. | | Deposit date: | 2020-12-22 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Active site architecture reveals coordination sphere flexibility and specificity determinants in a group of closely related molybdoenzymes.
J.Biol.Chem., 296, 2021
|
|
6U30
 
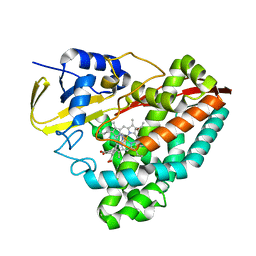 | | The crystal structure of 4-pyridin-3-ylbenzoate-bound CYP199A4 | | Descriptor: | 4-(pyridin-3-yl)benzoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Biophysical Techniques for Distinguishing Ligand Binding Modes in Cytochrome P450 Monooxygenases.
Biochemistry, 59, 2020
|
|
6U31
 
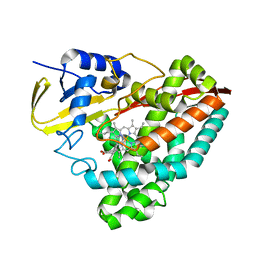 | | The crystal structure of 4-(1H-imidazol-1-yl)benzoate-bound CYP199A4 | | Descriptor: | 4-(1H-imidazol-1-yl)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-08-21 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.578 Å) | | Cite: | To Be, or Not to Be, an Inhibitor: A Comparison of Azole Interactions with and Oxidation by a Cytochrome P450 Enzyme.
Inorg.Chem., 61, 2022
|
|
6WZP
 
 | | The crystal structure of 4-vinylbenzoate-bound T252A mutant CYP199A4 | | Descriptor: | 4-ethenylbenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Coleman, T, Bruning, J.B, Bell, S.G. | | Deposit date: | 2020-05-14 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Understanding the Mechanistic Requirements for Efficient and Stereoselective Alkene Epoxidation by a Cytochrome P450 Enzyme
Acs Catalysis, 11, 2021
|
|
3OCD
 
 | | Diheme SoxAX - C236M mutant | | Descriptor: | HEME C, SoxA, SoxX | | Authors: | Maher, M.J. | | Deposit date: | 2010-08-09 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Diheme SoxAX proteins - insights into structure and function of the active site
To be Published
|
|
3OA8
 
 | | Diheme SoxAX | | Descriptor: | HEME C, SULFATE ION, SoxA, ... | | Authors: | Maher, M.J. | | Deposit date: | 2010-08-04 | | Release date: | 2011-05-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Diheme SoxAX proteins - insights into structure and function of the active site
To be Published
|
|
5K3X
 
 | | Crystal Structure of the sulfite dehydrogenase, SorT R78K mutant from Sinorhizobium meliloti | | Descriptor: | (MOLYBDOPTERIN-S,S)-OXO-MOLYBDENUM, GLYCEROL, Putative sulfite oxidase | | Authors: | Lee, M, McGrath, A, Maher, M. | | Deposit date: | 2016-05-20 | | Release date: | 2017-05-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The central active site arginine in sulfite oxidizing enzymes alters kinetic properties by controlling electron transfer and redox interactions.
Biochim. Biophys. Acta, 1859, 2017
|
|
7REH
 
 | |
8BAC
 
 | | Crystal structure of human heparanase in complex with competitive inhibitor GD05 | | Descriptor: | (3S,4R,5R)-4,5-dihydroxypiperidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Armstrong, Z, Davies, G.J. | | Deposit date: | 2022-10-11 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis of Uronic Acid 1-Azasugars as Putative Inhibitors of alpha-Iduronidase, beta-Glucuronidase and Heparanase.
Chembiochem, 24, 2023
|
|
2CA3
 
 | | Sulfite dehydrogenase from Starkeya Novella r55m mutant | | Descriptor: | (MOLYBDOPTERIN-S,S)-OXO-MOLYBDENUM, HEME C, SULFATE ION, ... | | Authors: | Bailey, S, Kappler, U, Feng, C, Honeychurch, M.J, Bernhardt, P.V, Tollin, G, Enemark, J.H. | | Deposit date: | 2005-12-16 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for enzymatic sulfite oxidation: how three conserved active site residues shape enzyme activity.
J.Biol.Chem., 284, 2009
|
|
