6GO1
 
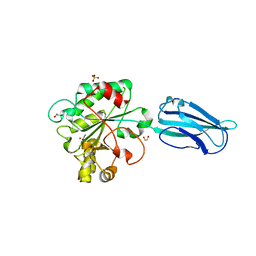 | | Crystal Structure of a Bacillus anthracis peptidoglycan deacetylase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Polysaccharide deacetylase-like protein, ... | | Authors: | Giastas, P, Andreou, A, Eliopoulos, E.E. | | Deposit date: | 2018-06-01 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The putative polysaccharide deacetylase Ba0331: cloning, expression, crystallization and structure determination.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5NEK
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, ACETATE ION, Peptidoglycan N-acetylglucosamine deacetylase, ... | | Authors: | Andreou, A, Giastas, P, Eliopoulos, E.E. | | Deposit date: | 2017-03-10 | | Release date: | 2018-02-21 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (3.057 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
5NEL
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with ThiametG | | Descriptor: | (3AR,5R,6S,7R,7AR)-2-(ETHYLAMINO)-5-(HYDROXYMETHYL)-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]THIAZOLE-6,7-DIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Andreou, A, Giastas, P, Eliopoulos, E.E. | | Deposit date: | 2017-03-10 | | Release date: | 2018-02-21 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
4V33
 
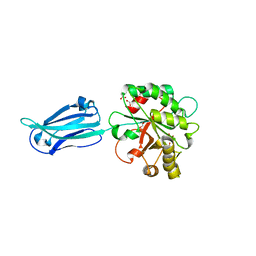 | | Crystal structure of the putative polysaccharide deacetylase BA0330 from bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, POLYSACCHARIDE DEACETYLASE-LIKE PROTEIN, ... | | Authors: | Giastas, P, Andreou, A, Bouriotis, V, Eliopoulos, E. | | Deposit date: | 2014-10-16 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Two Putative Polysaccharide Deacetylases are Required for Osmotic Stability and Cell Shape Maintenance in Bacillus Anthracis.
J.Biol.Chem., 290, 2015
|
|
5EK8
 
 | | Crystal structure of a 9R-lipoxygenase from Cyanothece PCC8801 at 2.7 Angstroms | | Descriptor: | FE (II) ION, Lipoxygenase, SODIUM ION | | Authors: | Feussner, I, Ficner, R, Neumann, P, Newie, J, Andreou, A, Einsle, O. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a lipoxygenase from Cyanothece sp. may reveal novel features for substrate acquisition.
J.Lipid Res., 57, 2016
|
|
5N1J
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Peptidoglycan N-acetylglucosamine deacetylase, ... | | Authors: | Giastas, P, Andreou, A, Balomenou, S, Bouriotis, V, Eliopoulos, E.E. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
5NC9
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with (2S)-2,6-diamino-N-hydroxyhexanamide | | Descriptor: | (2~{S})-2,6-bis(azanyl)-~{N}-oxidanyl-hexanamide, 1,2-ETHANEDIOL, CITRIC ACID, ... | | Authors: | Giastas, P, Andreou, A, Eliopoulos, E.E. | | Deposit date: | 2017-03-03 | | Release date: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
5N1P
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with N-hydroxynaphthalene-1-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, Peptidoglycan N-acetylglucosamine deacetylase, SODIUM ION, ... | | Authors: | Giastas, P, Andreou, A, Eliopoulos, E.E. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-21 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
5NC6
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with (E)-N-hydroxy-3-(naphthalen-1-yl)prop-2-enamide | | Descriptor: | 1,2-ETHANEDIOL, 3-naphthalen-1-yl-~{N}-oxidanyl-propanamide, ACETATE ION, ... | | Authors: | Giastas, P, Andreou, A, Balomenou, S, Bouriotis, V, Eliopoulos, E.E. | | Deposit date: | 2017-03-03 | | Release date: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
5NCD
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with (2S)-2-amino-5-(diaminomethylideneamino)-N-hydroxypentanamide | | Descriptor: | ACETATE ION, CITRIC ACID, N-HYDROXY-L-ARGININAMIDE, ... | | Authors: | Giastas, P, Andreou, A, Eliopoulos, E.E. | | Deposit date: | 2017-03-03 | | Release date: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
