3KDE
 
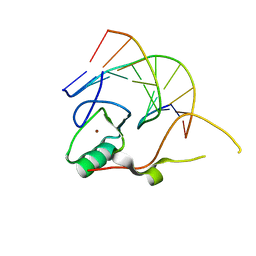 | | Crystal structure of the THAP domain from D. melanogaster P-element transposase in complex with its natural DNA binding site | | Descriptor: | 5'-D(*(BRU)P*CP*CP*AP*CP*TP*TP*AP*AP*C)-3', 5'-D(*GP*TP*TP*AP*AP*GP*(BRU)P*GP*GP*A)-3', Transposable element P transposase, ... | | Authors: | Sabogal, A, Lyubimov, A.Y, Berger, J.M, Rio, D.C. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | THAP proteins target specific DNA sites through bipartite recognition of adjacent major and minor grooves.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5JH0
 
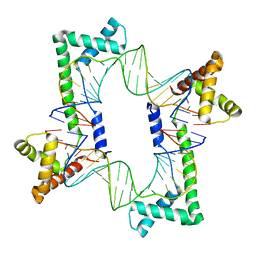 | | Crystal structure of the mitochondrial DNA packaging protein Abf2p in complex with DNA at 2.18 Angstrom resolution | | Descriptor: | ARS-binding factor 2, mitochondrial, DNA (5'-D(*AP*AP*TP*AP*AP*TP*AP*AP*AP*TP*TP*AP*TP*AP*TP*AP*AP*TP*AP*TP*AP*A)-3'), ... | | Authors: | Chakraborty, A, Lyonnais, S, Sola, M. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | DNA structure directs positioning of the mitochondrial genome packaging protein Abf2p.
Nucleic Acids Res., 45, 2017
|
|
5JGH
 
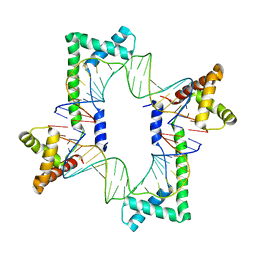 | | Crystal structure of the mitochondrial DNA packaging protein Abf2p in complex with DNA at 2.6 Angstrom resolution | | Descriptor: | ACETATE ION, ARS-binding factor 2, mitochondrial, ... | | Authors: | Chakraborty, A, Lyonnais, S, Sola, M. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DNA structure directs positioning of the mitochondrial genome packaging protein Abf2p.
Nucleic Acids Res., 45, 2017
|
|
7O3N
 
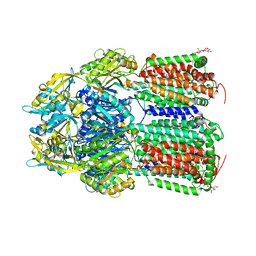 | | Crystal Structure of AcrB Single Mutant - 2 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Efflux pump membrane transporter | | Authors: | Ababou, A. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.561 Å) | | Cite: | Crystal Structure of AcrB Single Mutant - 2
To Be Published
|
|
7O3M
 
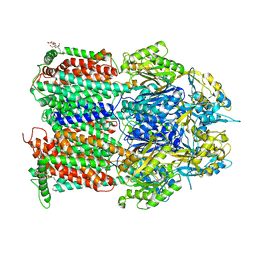 | | Crystal Structure of AcrB Single Mutant - 1 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Efflux pump membrane transporter | | Authors: | Ababou, A. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.551 Å) | | Cite: | Crystal Structure of AcrB Single Mutant - 1
To Be Published
|
|
7O3L
 
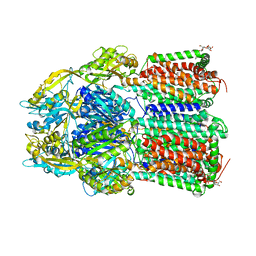 | | Crystal Structure of AcrB Double Mutant | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Efflux pump membrane transporter | | Authors: | Ababou, A. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.526 Å) | | Cite: | Crystal Structure of AcrB Double Mutant
To Be Published
|
|
4CRQ
 
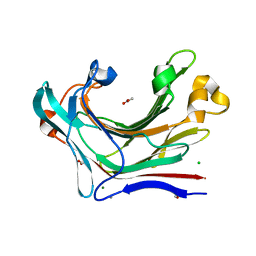 | | Crystal structure of the catalytic domain of the modular laminarinase ZgLamC mutant E142S | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Labourel, A, Jam, M, Legentil, L, Sylla, B, Ficko-Blean, E, Hehemann, J.H, Ferrieres, V, Czjzek, M, Michel, G. | | Deposit date: | 2014-02-28 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Biochemical Characterization of the Laminarina Zglamc[Gh16] from Zobellia Galactanivorans Suggests Preferred Recognition of Branched Laminarin
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4EZ2
 
 | |
5DSU
 
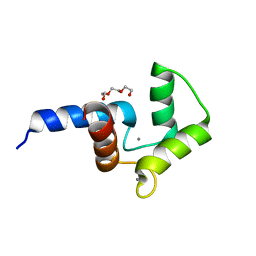 | | Crystal structure of double mutant of N-domain of human calmodulin | | Descriptor: | CALCIUM ION, Calmodulin, TRIETHYLENE GLYCOL | | Authors: | Ababou, A, Zaleska, M. | | Deposit date: | 2015-09-17 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | On the Ca(2+) binding and conformational change in EF-hand domains: Experimental evidence of Ca(2+)-saturated intermediates of N-domain of calmodulin.
Biochim. Biophys. Acta, 1865, 2017
|
|
4ZIT
 
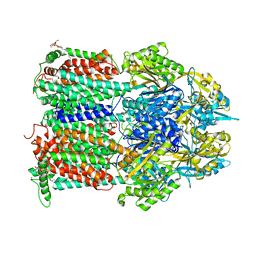 | | Crystal structure of AcrB in P21 space group | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Multidrug efflux pump subunit AcrB, NICKEL (II) ION | | Authors: | Ababou, A, Koronakis, V. | | Deposit date: | 2015-04-28 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.296 Å) | | Cite: | Structures of Gate Loop Variants of the AcrB Drug Efflux Pump Bound by Erythromycin Substrate.
Plos One, 11, 2016
|
|
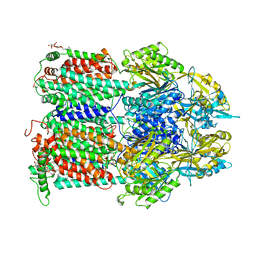
4ZJO
 
 | |
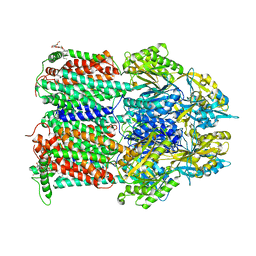
4ZIW
 
 | |
4ZLL
 
 | |
4ZLJ
 
 | |
4ZJQ
 
 | |
4ZJL
 
 | |
4ZLN
 
 | |
4ZIV
 
 | |
4BPZ
 
 | | Crystal structure of lamA_E269S from Zobellia galactanivorans in complex with a trisaccharide of 1,3-1,4-beta-D-glucan. | | Descriptor: | CALCIUM ION, ENDO-1,3-BETA-GLUCANASE, FAMILY GH16, ... | | Authors: | Labourel, A, Jam, M, Jeudy, A, Czjzek, M, Michel, G. | | Deposit date: | 2013-05-29 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | The Beta-Glucanase Zglama from Zobellia Galactanivorans Evolved a Bent Active Site Adapted for Efficient Degradation of Algal Laminarin.
J.Biol.Chem., 289, 2014
|
|
4BOW
 
 | | Crystal structure of LamA_E269S from Z. galactanivorans in complex with laminaritriose and laminaritetraose | | Descriptor: | CALCIUM ION, ENDO-1,3-BETA-GLUCANASE, FAMILY GH16, ... | | Authors: | Labourel, A, Jeudy, A, Czjzek, M, Michel, G. | | Deposit date: | 2013-05-22 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Beta-Glucanase Zglama from Zobellia Galactanivorans Evolved a Bent Active Site Adapted for Efficient Degradation of Algal Laminarin
J.Biol.Chem., 289, 2014
|
|
4BQ1
 
 | | Crystal structure of of LamAcat from Zobellia galactanivorans | | Descriptor: | CALCIUM ION, ENDO-1,3-BETA-GLUCANASE, FAMILY GH16, ... | | Authors: | Labourel, A, Jam, M, Jeudy, A, Michel, G, Czjzek, M. | | Deposit date: | 2013-05-29 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Beta Glucanase Zglama from Zobellia Galactanivorans Evolved a Bent Active Site Adapted for Efficient Degradation of Algal Laminarin
J.Biol.Chem., 289, 2014
|
|
1PD6
 
 | |
2AVG
 
 | |
2O2O
 
 | |
2I08
 
 | |
