1VTC
 
 | |
1VTW
 
 | | AT Base Pairs Are Less Stable than GC Base Pairs in Z-DNA: The Crystal Structure of D(M(5)CGTAM(5)CG) | | Descriptor: | DNA (5'-D(*(CH3)CP*GP*TP*AP*(CH3)CP*G)-3') | | Authors: | Wang, A.H.-J, Hakoshima, T, Van Der Marel, G.A, Van Boom, J.H, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | AT Base Pairs Are Less Stable than GC Base Pairs in Z-DNA: The Crystal Structure of d(m(5)CGTAm(5)CG)
Cell(Cambridge,Mass.), 37, 1984
|
|
2ZUG
 
 | | Crystal structure of WSSV ICP11 | | Descriptor: | ORF115 (WSSV285) (Wsv230) | | Authors: | Wang, A.H.-J, Wang, H.-C, Ko, T.-P, Lo, C.-F. | | Deposit date: | 2008-10-17 | | Release date: | 2008-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | White spot syndrome virus protein ICP11: A histone-binding DNA mimic that disrupts nucleosome assembly
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1VS2
 
 | | Interactions of quinoxaline antibiotic and DNA: the molecular structure of a TRIOSTIN A-D(GCGTACGC) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, 5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3', TRIOSTIN A | | Authors: | Wang, A.H.-J, Ughetto, G, Quigley, G.J, Rich, A. | | Deposit date: | 1986-10-21 | | Release date: | 2006-06-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interactions of Quinoxaline Antibiotic and DNA: The Molecular Structure of a Triostin A-D(Gcgtacgc) Complex.
J.Biomol.Struct.Dyn., 4, 1986
|
|
2DCG
 
 | | MOLECULAR STRUCTURE OF A LEFT-HANDED DOUBLE HELICAL DNA FRAGMENT AT ATOMIC RESOLUTION | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, SPERMINE | | Authors: | Wang, A.H.-J, Quigley, G.J, Kolpak, F.J, Crawford, J.L, Van Boom, J.H, Van Der Marel, G.A, Rich, A. | | Deposit date: | 1988-08-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Molecular structure of a left-handed double helical DNA fragment at atomic resolution.
Nature, 282, 1979
|
|
1CA5
 
 | | INTERCALATION SITE OF HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SSO7D/SAC7D BOUND TO DNA | | Descriptor: | 5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3', CHROMOSOMAL PROTEIN SAC7D | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1999-02-23 | | Release date: | 2000-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the chromosomal proteins Sso7d/Sac7d bound to DNA containing T-G mismatched base-pairs
J.Mol.Biol., 303, 2000
|
|
1CA6
 
 | | INTERCALATION SITE OF HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SSO7D/SAC7D BOUND TO DNA | | Descriptor: | 5'-D(*GP*TP*GP*AP*TP*CP*GP*C)-3', CHROMOSOMAL PROTEIN SAC7D | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1999-02-23 | | Release date: | 2000-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the chromosomal proteins Sso7d/Sac7d bound to DNA containing T-G mismatched base-pairs
J.Mol.Biol., 303, 2000
|
|
5G50
 
 | |
1MNV
 
 | | Actinomycin D binding to ATGCTGCAT | | Descriptor: | 5'-D(*AP*TP*GP*CP*TP*GP*CP*AP*T)-3', ACTINOMYCIN D | | Authors: | Hou, M.-H, Robinson, H, Gao, Y.-G, Wang, A.H.-J. | | Deposit date: | 2002-09-06 | | Release date: | 2002-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Actinomycin D Bound to the Ctg Triplet Repeat Sequences Linked to Neurological Diseases
Nucleic Acids Res., 30, 2002
|
|
1NDN
 
 | | MOLECULAR STRUCTURE OF NICKED DNA. MODEL T4 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*AP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*T)-3'), DNA (5'-D(*TP*TP*CP*GP*CP*G)-3') | | Authors: | Aymani, J, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1992-01-15 | | Release date: | 1992-07-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular structure of nicked DNA: a substrate for DNA repair enzymes.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
4EJT
 
 | |
2L66
 
 | |
3MBR
 
 | | Crystal Structure of the Glutaminyl Cyclase from Xanthomonas campestris | | Descriptor: | CALCIUM ION, Glutamine cyclotransferase | | Authors: | Huang, W.-L, Wang, Y.-R, Ko, T.-P, Chia, C.-Y, Huang, K.-F, Wang, A.H.-J. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structure and functional analysis of the glutaminyl cyclase from Xanthomonas campestris
J.Mol.Biol., 401, 2010
|
|
3O98
 
 | | Glutathionylspermidine synthetase/amidase C59A complex with ADP and Gsp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional glutathionylspermidine synthetase/amidase, GLUTATHIONYLSPERMIDINE, ... | | Authors: | Pai, C.H, Lin, C.H, Wang, A.H.-J. | | Deposit date: | 2010-08-04 | | Release date: | 2011-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of Escherichia coli glutathionylspermidine amidase belonging to the family of cysteine; histidine-dependent amidohydrolases/peptidases
Protein Sci., 20, 2011
|
|
1WL2
 
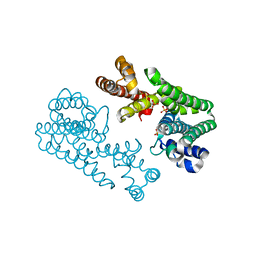 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima R90A mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
1WL1
 
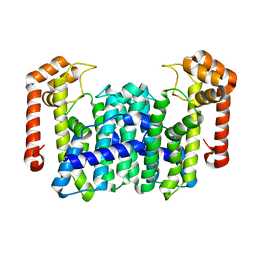 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima H74A mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
1WL0
 
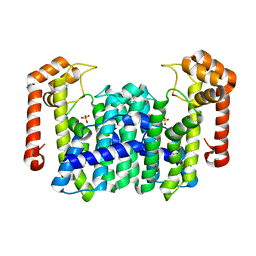 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima R44A mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
1WL3
 
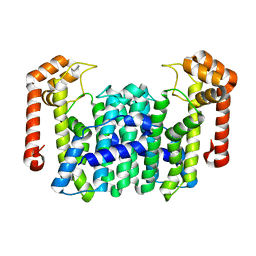 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima R91A mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
1WKZ
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima K41A mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
1CYZ
 
 | | NMR STRUCTURE OF THE GAACTGGTTC/TRI-IMIDAZOLE POLYAMIDE COMPLEX | | Descriptor: | (2-{[4-({4-[(4-FORMYLAMINO-1-METHYL-1H-IMIDAZOLE-2-CARBONYL)-AMINO]-1-METHYL-1H-IMIDAZOLE-2-CARBONYL}-AMINO)-1-METHYL-1 H-IMIDAZOLE-2-CARBONYL]-AMINO}-ETHYL)-DIMETHYL-AMMONIUM, 5'-D(*GP*AP*AP*CP*TP*GP*GP*TP*TP*C)-3' | | Authors: | Yang, X.-L, Hubbard IV, R.B, Lee, M, Tao, Z.-F, Sugiyama, H, Wang, A.H.-J. | | Deposit date: | 1999-08-31 | | Release date: | 1999-09-14 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Imidazole-imidazole pair as a minor groove recognition motif for T:G mismatched base pairs
Nucleic Acids Res., 27, 1999
|
|
1D22
 
 | | BINDING OF THE ANTITUMOR DRUG NOGALAMYCIN AND ITS DERIVATIVES TO DNA: STRUCTURAL COMPARISON | | Descriptor: | DNA (5'-D(*(5CM)P*GP*TP*(AS)P*(5CM)P*G)-3'), U-58872, HYDROXY DERIVATIVE OF NOGALAMYCIN | | Authors: | Gao, Y.-G, Liaw, Y.-C, Robinson, H, Wang, A.H.-J. | | Deposit date: | 1990-08-08 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of the antitumor drug nogalamycin and its derivatives to DNA: structural comparison.
Biochemistry, 29, 1990
|
|
1D21
 
 | | BINDING OF THE ANTITUMOR DRUG NOGALAMYCIN AND ITS DERIVATIVES TO DNA: STRUCTURAL COMPARISON | | Descriptor: | DNA (5'-D(*(5CM)P*GP*TP*(AS)P*(5CM)P*G)-3'), NOGALAMYCIN | | Authors: | Gao, Y.-G, Liaw, Y.-C, Robinson, H, Wang, A.H.-J. | | Deposit date: | 1990-08-08 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of the antitumor drug nogalamycin and its derivatives to DNA: structural comparison.
Biochemistry, 29, 1990
|
|
1D39
 
 | | COVALENT MODIFICATION OF GUANINE BASES IN DOUBLE STRANDED DNA: THE 1.2 ANGSTROMS Z-DNA STRUCTURE OF D(CGCGCG) IN THE PRESENCE OF CUCL2 | | Descriptor: | COPPER (II) ION, DNA (5'-D(*CP*(CU)GP*CP*(CU)GP*CP*(CU)G)-3'), SODIUM ION | | Authors: | Kagawa, T.F, Geierstanger, B.H, Wang, A.H.-J, Ho, P.S. | | Deposit date: | 1991-05-07 | | Release date: | 1992-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Covalent modification of guanine bases in double-stranded DNA. The 1.2-A Z-DNA structure of d(CGCGCG) in the presence of CuCl2.
J.Biol.Chem., 266, 1991
|
|
4BE6
 
 | |
4EM1
 
 | |
