1OHM
 
 | | Sakacin P variant that is structurally stabilized by an inserted C-terminal disulfide bridge. | | Descriptor: | BACTERIOCIN SAKACIN P | | Authors: | Uteng, M, Hauge, H.H, Markwick, P.R, Fimland, G, Mantzilas, D, Nissen-Meyer, J, Muhle-Goll, C. | | Deposit date: | 2003-05-28 | | Release date: | 2003-09-22 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure in Lipid Micelles of the Pediocin-Like Antimicrobial Peptide Sakacin P and a Sakacin P Variant that is Structurally Stabilized by an Inserted C-Terminal Disulfide Bridge
Biochemistry, 42, 2003
|
|
1OHN
 
 | | Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide sakacin P. | | Descriptor: | BACTERIOCIN SAKACIN P | | Authors: | Uteng, M, Hauge, H.H, Markwick, P.R, Fimland, G, Mantzilas, D, Nissen-Meyer, J, Muhle-Goll, C. | | Deposit date: | 2003-05-28 | | Release date: | 2003-09-22 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure in Lipid Micelles of the Pediocin-Like Antimicrobial Peptide Sakacin P and a Sakacin P Variant that is Structurally Stabilized by an Inserted C-Terminal Disulfide Bridge
Biochemistry, 42, 2003
|
|
1OG7
 
 | | Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide sakacin P. | | Descriptor: | BACTERIOCIN SAKACIN P | | Authors: | Uteng, M, Hauge, H.H, Markwick, P.R, Fimland, G, Mantzilas, D, Nissen-Meyer, J, Muhle-Goll, C. | | Deposit date: | 2003-04-25 | | Release date: | 2003-09-22 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure in Lipid Micelles of the Pediocin-Like Antimicrobial Peptide Sakacin P and a Sakacin P Variant that is Structurally Stabilized by an Inserted C-Terminal Disulfide Bridge
Biochemistry, 42, 2003
|
|
4KDC
 
 | | Crystal Structure of UBIG | | Descriptor: | 3-demethylubiquinone-9 3-methyltransferase | | Authors: | Zhu, Y, Teng, M, Li, X. | | Deposit date: | 2013-04-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and biochemical studies reveal UbiG/Coq3 as a class of novel membrane-binding proteins.
Biochem. J., 470, 2015
|
|
4KR1
 
 | |
1TDN
 
 | |
1TDO
 
 | |
1TDK
 
 | |
1REO
 
 | | L-amino acid oxidase from Agkistrodon halys pallas | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AHPLAAO, CITRIC ACID, ... | | Authors: | Zhang, H, Teng, M, Niu, L, Wang, Y, Wang, Y, Liu, Q, Huang, Q, Hao, Q, Dong, Y, Liu, P. | | Deposit date: | 2003-11-07 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Purification, partial characterization, crystallization and structural determination of AHP-LAAO, a novel L-amino-acid oxidase with cell apoptosis-inducing activity from Agkistrodon halys pallas venom.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OP2
 
 | | Crystal Structure of AaV-SP-II, a Glycosylated Snake Venom Serine Proteinase from Agkistrodon acutus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Venom serine proteinase | | Authors: | Zhu, Z, Teng, M, Niu, L. | | Deposit date: | 2003-03-04 | | Release date: | 2004-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures and Amidolytic Activities of Two Glycosylated Snake Venom Serine Proteinases
J.BIOL.CHEM., 280, 2005
|
|
1OP0
 
 | | Crystal Structure of AaV-SP-I, a Glycosylated Snake Venom Serine Proteinase from Agkistrodon acutus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Venom serine proteinase | | Authors: | Zhu, Z, Teng, M, Niu, L. | | Deposit date: | 2003-03-04 | | Release date: | 2004-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures and Amidolytic Activities of Two Glycosylated Snake Venom Serine Proteinases
J.BIOL.CHEM., 280, 2005
|
|
1QT1
 
 | |
1QUA
 
 | | CRYSTAL STRUCTURE OF ACUTOLYSIN-C, A HEMORRHAGIC TOXIN FROM THE SNAKE VENOM OF AGKISTRODON ACUTUS, AT 2.2 A RESOLUTION | | Descriptor: | ACUTOLYSIN-C, ZINC ION | | Authors: | Niu, L, Teng, M, Zhu, X. | | Deposit date: | 1999-06-30 | | Release date: | 2000-07-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of acutolysin-C, a haemorrhagic toxin from the venom of Agkistrodon acutus, providing further evidence for the mechanism of the pH-dependent proteolytic reaction of zinc metalloproteinases.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BSW
 
 | | ACUTOLYSIN A FROM SNAKE VENOM OF AGKISTRODON ACUTUS AT PH 7.5 | | Descriptor: | CALCIUM ION, PROTEIN (ACUTOLYSIN A), ZINC ION | | Authors: | Gong, W, Zhu, X, Liu, S, Teng, M, Niu, L. | | Deposit date: | 1998-08-31 | | Release date: | 1999-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of acutolysin A, a three-disulfide hemorrhagic zinc metalloproteinase from the snake venom of Agkistrodon acutus.
J.Mol.Biol., 283, 1998
|
|
1CLK
 
 | | CRYSTAL STRUCTURE OF STREPTOMYCES DIASTATICUS NO.7 STRAIN M1033 XYLOSE ISOMERASE AT 1.9 A RESOLUTION WITH PSEUDO-I222 SPACE GROUP | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Niu, L, Teng, M, Zhu, X, Gong, W. | | Deposit date: | 1999-04-29 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of xylose isomerase from Streptomyces diastaticus no. 7 strain M1033 at 1.85 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1W24
 
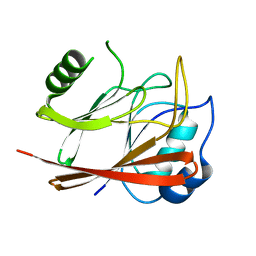 | | Crystal Structure Of human Vps29 | | Descriptor: | VACUOLAR PROTEIN SORTING PROTEIN 29 | | Authors: | Wang, D, Guo, M, Teng, M, Niu, L. | | Deposit date: | 2004-06-26 | | Release date: | 2005-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Vacuolar Protein Sorting Protein 29 Reveals a Phosphodiesterase/Nuclease-Like Fold and Two Protein-Protein Interaction Sites.
J.Biol.Chem., 280, 2005
|
|
1BUD
 
 | | ACUTOLYSIN A FROM SNAKE VENOM OF AGKISTRODON ACUTUS AT PH 5.0 | | Descriptor: | CALCIUM ION, PROTEIN (ACUTOLYSIN A), ZINC ION | | Authors: | Gong, W, Zhu, X, Liu, S, Teng, M, Niu, L. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of acutolysin A, a three-disulfide hemorrhagic zinc metalloproteinase from the snake venom of Agkistrodon acutus.
J.Mol.Biol., 283, 1998
|
|
5DPM
 
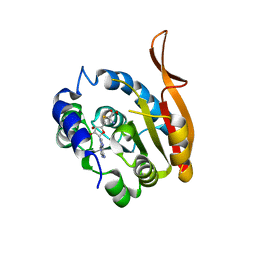 | |
1MG6
 
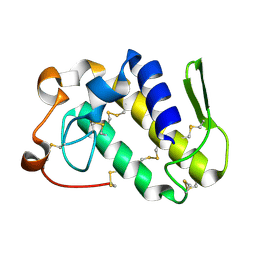 | |
1ZVF
 
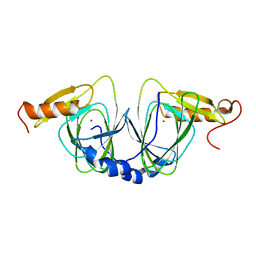 | | The crystal structure of 3-hydroxyanthranilate 3,4-dioxygenase from Saccharomyces cerevisiae | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, NICKEL (II) ION | | Authors: | Li, X, Guo, M, Teng, M, Niu, L. | | Deposit date: | 2005-06-02 | | Release date: | 2006-06-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The crystal structure of 3-hydroxyanthranilate 3,4-dioxygenase from Saccharomyces cerevisiae
To be published
|
|
5BT1
 
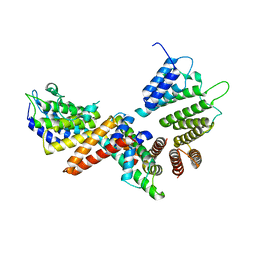 | | histone chaperone Hif1 playing with histone H2A-H2B dimer | | Descriptor: | HAT1-interacting factor 1, Histone H2A.1, Histone H2B.1 | | Authors: | Liu, H, Zhang, M, Gao, Y, Teng, M, Niu, L. | | Deposit date: | 2015-06-02 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural Insights into the Association of Hif1 with Histones H2A-H2B Dimer and H3-H4 Tetramer
Structure, 24, 2016
|
|
1VB0
 
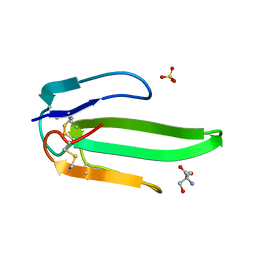 | | Atomic resolution structure of atratoxin-b, one short-chain neurotoxin from Naja atra | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cobrotoxin b, SULFATE ION | | Authors: | Lou, X, Liu, Q, Teng, M, Niu, L, Huang, Q, Hao, Q. | | Deposit date: | 2004-02-20 | | Release date: | 2004-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | The atomic resolution crystal structure of atratoxin determined by single wavelength anomalous diffraction phasing
J.Biol.Chem., 279, 2004
|
|
2EXU
 
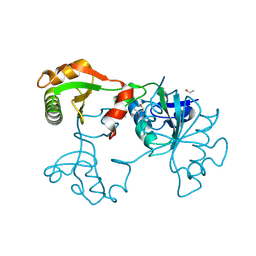 | | Crystal Structure of Saccharomyces cerevisiae transcription elongation factors Spt4-Spt5NGN domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ETHANOL, Transcription initiation protein SPT4/SPT5, ... | | Authors: | Xu, F, Guo, M, Fang, P, Teng, M, Niu, L. | | Deposit date: | 2005-11-08 | | Release date: | 2006-11-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal Structure of Saccharomyces cerevisiae transcription elongation factors Spt4-Spt5NGN domain
To be published
|
|
2FQD
 
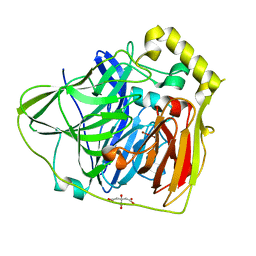 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2FQE
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
