5YQW
 
 | | Structure and function of a novel periplasmic chitooligosaccharide-binding protein from marine Vibrio bacteria | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NICKEL (II) ION, ... | | Authors: | Suginta, W, Sritho, N, Ranok, A, Kitaoku, Y, Bulmer, D.M, van den Berg, B, Fukamizo, T. | | Deposit date: | 2017-11-08 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure and function of a novel periplasmic chitooligosaccharide-binding protein from marineVibriobacteria.
J. Biol. Chem., 293, 2018
|
|
7VU0
 
 | | Chitoporin from Escherichia coli | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Chitoporin, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Suginta, W, Soysa, H.S.M, Amornloetwattana, R, van den Berg, B. | | Deposit date: | 2021-11-01 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Chitoporin from Escherichia coli
To Be Published
|
|
4P8V
 
 | | The crystal structures of YKL-39 in the presence of chitooligosaccharides (GlcNAc2) were solved to resolutions of 1.5 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 2, SULFATE ION | | Authors: | Suginta, W, Ranok, A, Robinson, R.C, Wongsantichon, J. | | Deposit date: | 2014-04-01 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural and Thermodynamic Insights into Chitooligosaccharide Binding to Human Cartilage Chitinase 3-like Protein 2 (CHI3L2 or YKL-39).
J.Biol.Chem., 290, 2015
|
|
4P8X
 
 | | The crystal structures of YKL-39 in the presence of chitooligosaccharides (GlcNAc6) were solved to resolutions of 2.48 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 2, SULFATE ION | | Authors: | Suginta, W, Ranok, A, Robinson, R.C, Wongsantichon, J. | | Deposit date: | 2014-04-01 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural and Thermodynamic Insights into Chitooligosaccharide Binding to Human Cartilage Chitinase 3-like Protein 2 (CHI3L2 or YKL-39).
J.Biol.Chem., 290, 2015
|
|
4P8U
 
 | | The crystal structures of YKL-39 in the absence of chitooligosaccharides was solved to resolutions of 2.4 angstrom | | Descriptor: | Chitinase-3-like protein 2, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Suginta, W, Ranok, A, Robinson, R.C, Wongsantichon, J. | | Deposit date: | 2014-04-01 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Thermodynamic Insights into Chitooligosaccharide Binding to Human Cartilage Chitinase 3-like Protein 2 (CHI3L2 or YKL-39).
J.Biol.Chem., 290, 2015
|
|
4P8W
 
 | | The crystal structures of YKL-39 in the presence of chitooligosaccharides (GlcNAc4) were solved to resolutions of 1.9 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase-3-like protein 2, SULFATE ION | | Authors: | Suginta, W, Ranok, A, Robinson, R.C, Wongsantichon, J. | | Deposit date: | 2014-04-01 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Thermodynamic Insights into Chitooligosaccharide Binding to Human Cartilage Chitinase 3-like Protein 2 (CHI3L2 or YKL-39).
J.Biol.Chem., 290, 2015
|
|
7VU1
 
 | | Chitoporin from Escherichia coli complex with chitohexaose | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitoporin, ... | | Authors: | Suginta, W, Soysa, H.S.M, Amornloetwattana, R, van den Berg, B. | | Deposit date: | 2021-11-01 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chitporin from Escherichia coli complex with chitohexaose
To Be Published
|
|
6EZT
 
 | | Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase D437A inactive mutant from Vibrio harveyi | | Descriptor: | Beta-N-acetylglucosaminidase Nag2, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL | | Authors: | Porfetye, A.T, Meekrathok, P, Burger, M, Vetter, I.R, Suginta, W. | | Deposit date: | 2017-11-16 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase from Vibrio harveyi
To Be Published
|
|
6EZR
 
 | |
6EZS
 
 | | Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase from Vibrio harveyi in complex with N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylglucosaminidase Nag2, MALONATE ION | | Authors: | Porfetye, A.T, Meekrathok, P, Burger, M, Vetter, I.R, Suginta, W. | | Deposit date: | 2017-11-16 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase from Vibrio harveyi
To Be Published
|
|
4GNJ
 
 | | Crystal Structure Analysis of Leishmania siamensis Triosephosphate Isomerase | | Descriptor: | ARSENIC, SODIUM ION, Triosephosphate isomerase | | Authors: | Kuaprasert, B, Riangrungroj, P, Pornthanakasem, W, Suginta, W, Mungthin, M, Leelayoova, S, Leartsakulpanich, U. | | Deposit date: | 2012-08-17 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure Analysis of Leishmania siamensis Triosephosphate Isomerase
To be Published
|
|
8GUL
 
 | |
8GUM
 
 | |
8J7C
 
 | | Crystal structure of triosephosphate isomerase from Leishmania orientalis at 1.88A with an arsenic ion bound at Cys57 | | Descriptor: | ARSENIC, Triosephosphate isomerase | | Authors: | Kuaprasert, B, Attarataya, J, Riangrungroj, P, Pornthanakasem, W, Suginta, W, Mungthin, M, Leelayoova, S, Choowongkomon, K, Leartsakulpanich, U. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Leishmania orientalis triosephosphate isomerase crystal structure at 1.45 angstroms resolution and its potential specific inhibitors
To be published
|
|
8JBP
 
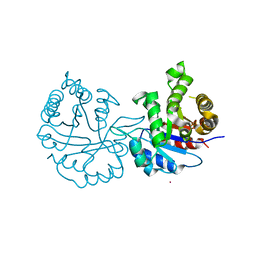 | | Crystal structure of triosephosphate isomerase from Leishmania orientalis at 1.45 angstroms resolution with an arsenic atom bound at Cys57 | | Descriptor: | ARSENIC, Triosephosphate isomerase | | Authors: | Kuaprasert, B, Leartsakulpanich, U, Riangrungroj, P, Pornthanakasem, W, Suginta, W, Robinson, R.C, Zhou, Y, Mungthin, M, Leelayoova, S, Saehlee, S, Choowongkomon, K. | | Deposit date: | 2023-05-09 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Leishmania orientalis triosephosphate isomerase crystal structure at 1.45 angstroms resolution and its potential specific inhibitors
To be published
|
|
8JTO
 
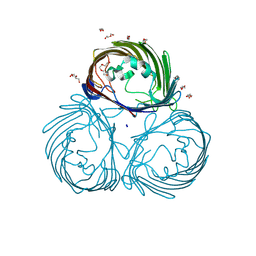 | | Outer membrane porin of Burkholderia pseudomallei (BpsOmp38) in complex with ceftazidime | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Outer membrane porin (Fragment), SODIUM ION | | Authors: | Bunkum, P, Aunkham, A, Bert van den, B, Robinson, R.C, Suginta, W. | | Deposit date: | 2023-06-22 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and function of outer membrane protein from Burkholderia pseudomallei (BpsOmp38)
To be published
|
|
7EQM
 
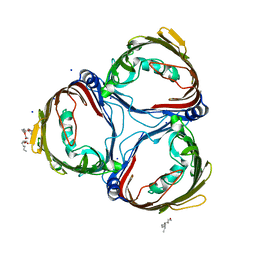 | | Apo Truncated VhChiP (Delta 1-19) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Chitoporin, MAGNESIUM ION, ... | | Authors: | Aunkham, A, Sanram, S, Suginta, W. | | Deposit date: | 2021-05-04 | | Release date: | 2022-08-10 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.50002384 Å) | | Cite: | Structural displacement model of chitooligosaccharide transport through chitoporin.
J.Biol.Chem., 299, 2023
|
|
8JUR
 
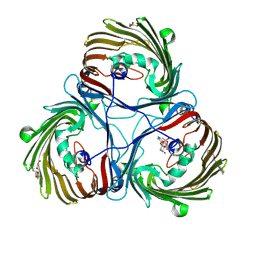 | | Crystal structure of Chitoporin from Vibrio harveyi in complex with gentamicin c1a (multiple binding sites) | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Chitoporin, ... | | Authors: | Sanram, S, Suginta, W, Robinson, R. | | Deposit date: | 2023-06-27 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Crystal structure of Chitoporin from Vibrio harveyi in complex with gentamicin c1a (multiple binding sites)
To Be Published
|
|
8JUQ
 
 | | Crystal structure of Chitoporin from Vibrio harveyi in complex with gentamicin c1a | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Chitoporin, ... | | Authors: | Sanram, S, Suginta, W, Robinson, R. | | Deposit date: | 2023-06-27 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of Chitoporin from Vibrio harveyi in complex with gentamicin c1a
To Be Published
|
|
7EBM
 
 | | W363A mutant of Chitin-specific solute binding protein from Vibrio harveyi in complex with chitobiose. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kitaoku, Y, Ubonbal, P, Tran, L.T, Robinson, R.C, Suginta, W. | | Deposit date: | 2021-03-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural model for (GlcNAc) 2 translocation via a periplasmic chitooligosaccharide-binding protein from marine Vibrio bacteria.
J.Biol.Chem., 297, 2021
|
|
7EBI
 
 | | Chitin-specific solute binding protein from Vibrio harveyi co-crystalized with chitotetraose. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kitaoku, Y, Ubonbal, P, Tran, L.T, Robinson, R.C, Suginta, W. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structural model for (GlcNAc) 2 translocation via a periplasmic chitooligosaccharide-binding protein from marine Vibrio bacteria.
J.Biol.Chem., 297, 2021
|
|
3ARQ
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with IDARUBICIN | | Descriptor: | Chitinase A, GLYCEROL, IDARUBICIN | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3AS3
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - W275G mutant complex structure with 2-(imidazolin-2-yl)-5-isothiocyanatobenzofuran | | Descriptor: | 2-(5-isothiocyanato-1-benzofuran-2-yl)-4,5-dihydro-1H-imidazole, Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3ARV
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with Sanguinarine | | Descriptor: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3ARZ
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with 2-(imidazolin-2-yl)-5-isothiocyanatobenzofuran | | Descriptor: | 2-(5-isothiocyanato-1-benzofuran-2-yl)-4,5-dihydro-1H-imidazole, Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
