3GYX
 
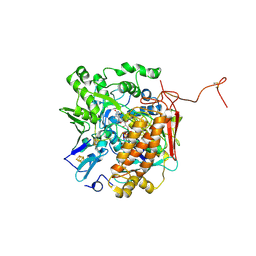 | | Crystal structure of adenylylsulfate reductase from Desulfovibrio gigas | | Descriptor: | Adenylylsulfate Reductase, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER | | Authors: | Chiang, Y.-L, Hsieh, Y.-C, Liu, E.-H, Liu, M.-Y, Chen, C.-J. | | Deposit date: | 2009-04-06 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Adenylylsulfate reductase from Desulfovibrio gigas suggests a potential self-regulation mechanism involving the C terminus of the beta-subunit
J.Bacteriol., 191, 2009
|
|
1NB2
 
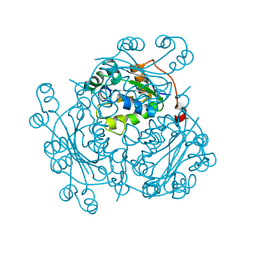 | | Crystal Structure of Nucleoside Diphosphate Kinase from Bacillus Halodenitrificans | | Descriptor: | Nucleoside Diphosphate Kinase | | Authors: | Chen, C.-J, Liu, M.-Y, Chang, W.-C, Chang, T, Wang, B.-C, Le Gall, J. | | Deposit date: | 2002-12-02 | | Release date: | 2003-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a nucleoside diphosphate kinase from Bacillus halodenitrificans: coexpression of its activity with a Mn-superoxide dismutase.
J.Struct.Biol., 142, 2003
|
|
1E5D
 
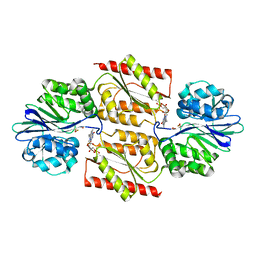 | | RUBREDOXIN OXYGEN:OXIDOREDUCTASE (ROO) FROM ANAEROBE DESULFOVIBRIO GIGAS | | Descriptor: | FLAVIN MONONUCLEOTIDE, MU-OXO-DIIRON, OXYGEN MOLECULE, ... | | Authors: | Frazao, C, Silva, G, Gomes, C.M, Matias, P, Coelho, R, Sieker, L, Macedo, S, Liu, M.Y, Oliveira, S, Teixeira, M, Xavier, A.V, Rodrigues-Pousada, C, Carrondo, M.A, Le Gall, J. | | Deposit date: | 2000-07-24 | | Release date: | 2000-11-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Dioxygen Reduction Enzyme from Desulfovibrio Gigas
Nat.Struct.Biol., 7, 2000
|
|
2NRD
 
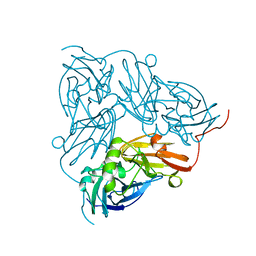 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1B71
 
 | | RUBRERYTHRIN | | Descriptor: | FE (III) ION, PROTEIN (RUBRERYTHRIN), ZINC ION | | Authors: | Sieker, L.C, Holmes, M, Le Trong, I, Turley, S, Santarsiero, B.D, Liu, M.-Y, Legall, J, Stenkamp, R.E. | | Deposit date: | 1999-01-26 | | Release date: | 2000-01-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternative metal-binding sites in rubrerythrin.
Nat.Struct.Biol., 6, 1999
|
|
1NIF
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1NIB
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1NIE
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
2DSX
 
 | | Crystal structure of rubredoxin from Desulfovibrio gigas to ultra-high 0.68 A resolution | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Chen, C.-J, Lin, Y.-H, Huang, Y.-C, Liu, M.-Y. | | Deposit date: | 2006-07-07 | | Release date: | 2006-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.68 Å) | | Cite: | Crystal structure of rubredoxin from Desulfovibrio gigas to ultra-high 0.68A resolution
Biochem.Biophys.Res.Commun., 349, 2006
|
|
1NID
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1NIA
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1NIC
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE, SULFATE ION | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1E8J
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS ZINC RUBREDOXIN, NMR, 20 STRUCTURES | | Descriptor: | RUBREDOXIN | | Authors: | Lamosa, P, Brennan, L, Vis, H, Turner, D.L, Santos, H. | | Deposit date: | 2000-09-21 | | Release date: | 2001-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Desulfovibrio gigas rubredoxin: a model for studying protein stabilization by compatible solutes.
Extremophiles, 5, 2001
|
|
1VLB
 
 | | STRUCTURE REFINEMENT OF THE ALDEHYDE OXIDOREDUCTASE FROM DESULFOVIBRIO GIGAS AT 1.28 A | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ALDEHYDE OXIDOREDUCTASE, CHLORIDE ION, ... | | Authors: | Rebelo, J.M, Dias, J.M, Huber, R, Moura, J.J.G, Romao, M.J. | | Deposit date: | 2004-07-20 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure refinement of the aldehyde oxidoreductase from Desulfovibrio gigas (MOP) at 1.28 A
J.Biol.Inorg.Chem., 6, 2001
|
|
