4P1M
 
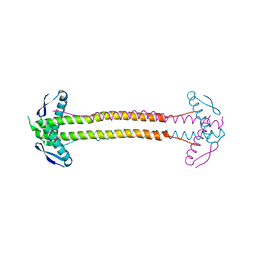 | | The structure of Escherichia coli ZapA | | Descriptor: | CHLORIDE ION, Cell division protein ZapA | | Authors: | Kimber, M.S, Roach, E.J, Khursigara, C.M. | | Deposit date: | 2014-02-26 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure and Site-directed Mutational Analysis Reveals Key Residues Involved in Escherichia coli ZapA Function.
J.Biol.Chem., 289, 2014
|
|
7SA8
 
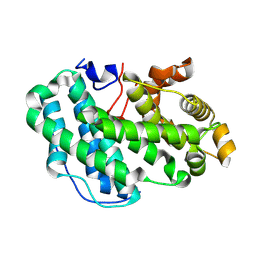 | |
2GRX
 
 | | Crystal structure of TonB in complex with FhuA, E. coli outer membrane receptor for ferrichrome | | Descriptor: | 2-AMINO-VINYL-PHOSPHATE, 3-HYDROXY-TETRADECANOIC ACID, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-[L-glycero-alpha-D-manno-heptopyranose-(1-5)]3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2,3-dideoxy-alpha-D-glucoyranose-(1-6)-2-amino-2,3-dideoxy-alpha-D-glucoyranose, ... | | Authors: | Pawelek, P.D, Allaire, M, Coulton, J.W. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of TonB in complex with FhuA, E. coli outer membrane receptor.
Science, 312, 2006
|
|
5DKO
 
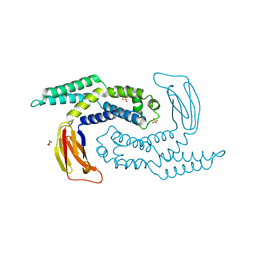 | | The structure of Escherichia coli ZapD | | Descriptor: | Cell division protein ZapD, SULFATE ION | | Authors: | Wroblewski, C, Kimber, M.S. | | Deposit date: | 2015-09-03 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Mutational Analyses of Escherichia coli ZapD Reveal Charged Residues Involved in FtsZ Filament Bundling.
J.Bacteriol., 198, 2016
|
|
4OZV
 
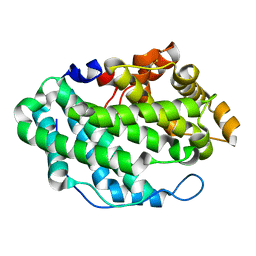 | | Crystal Structure of the periplasmic alginate lyase AlgL | | Descriptor: | Alginate lyase, beta-D-mannopyranuronic acid | | Authors: | Howell, P.L, Wolfram, F, Robinson, H, Arora, K. | | Deposit date: | 2014-02-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | The Pseudomonas aeruginosa homeostasis enzyme AlgL clears the periplasmic space of accumulated alginate during polymer biosynthesis.
J.Biol.Chem., 298, 2022
|
|
4OZW
 
 | |
