1GYN
 
 | | Class II fructose 1,6-bisphosphate aldolase with Cadmium (not Zinc) in the active site | | Descriptor: | CADMIUM ION, FRUCTOSE-BISPHOSPHATE ALDOLASE II | | Authors: | Hall, D.R, Kemp, L.E, Leonard, G.A, Berry, A, Hunter, W.N. | | Deposit date: | 2002-04-27 | | Release date: | 2003-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Organization of Divalent Cations in the Active Site of Cadmium Escherichia Coli Fructose 1,6-Bisphosphate Aldolase
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1GVF
 
 | | Structure of tagatose-1,6-bisphosphate aldolase | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOGLYCOLOHYDROXAMIC ACID, SODIUM ION, ... | | Authors: | Hall, D.R, Hunter, W.N. | | Deposit date: | 2002-02-11 | | Release date: | 2002-06-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of Tagatose-1,6-Bisphosphate Aldolase; Insight Into Chiral Discrimination, Mechanism and Specificity of Class II Aldolases
J.Biol.Chem., 277, 2002
|
|
1B9M
 
 | | REGULATOR FROM ESCHERICHIA COLI | | Descriptor: | NICKEL (II) ION, PROTEIN (MODE) | | Authors: | Hall, D.R, Gourley, D.G, Hunter, W.N. | | Deposit date: | 1999-02-12 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The high-resolution crystal structure of the molybdate-dependent transcriptional regulator (ModE) from Escherichia coli: a novel combination of domain folds.
EMBO J., 18, 1999
|
|
1B9N
 
 | | REGULATOR FROM ESCHERICHIA COLI | | Descriptor: | NICKEL (II) ION, PROTEIN (MODE) | | Authors: | Hall, D.R, Gourley, D.G, Hunter, W.N. | | Deposit date: | 1999-02-12 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The high-resolution crystal structure of the molybdate-dependent transcriptional regulator (ModE) from Escherichia coli: a novel combination of domain folds.
EMBO J., 18, 1999
|
|
1B57
 
 | | CLASS II FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE IN COMPLEX WITH PHOSPHOGLYCOLOHYDROXAMATE | | Descriptor: | CHLORIDE ION, PHOSPHOGLYCOLOHYDROXAMIC ACID, PROTEIN (FRUCTOSE-BISPHOSPHATE ALDOLASE II), ... | | Authors: | Hall, D.R, Hunter, W.N. | | Deposit date: | 1999-01-12 | | Release date: | 2000-01-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Escherichia coli class II fructose-1, 6-bisphosphate aldolase in complex with phosphoglycolohydroxamate reveals details of mechanism and specificity.
J.Mol.Biol., 287, 1999
|
|
1JNF
 
 | | Rabbit serum transferrin at 2.6 A resolution. | | Descriptor: | CARBONATE ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Hall, D.R, Hadden, J.M, Leonard, G.A, Bailey, S, Neu, M, Winn, M, Lindley, P.F. | | Deposit date: | 2001-07-24 | | Release date: | 2001-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal and molecular structures of diferric porcine and rabbit serum transferrins at resolutions of 2.15 and 2.60 A, respectively.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1H76
 
 | | The crystal structure of diferric porcine serum transferrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Hall, D.R, Hadden, J.M, Leonard, G.A, Bailey, S, Neu, M, Winn, M, Lindley, P.F. | | Deposit date: | 2001-07-03 | | Release date: | 2002-01-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal and Molecular Structures of Diferric Porcine and Rabbit Serum Transferrins at Resolutions of 2.15 And 2.60A, Respectively
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1QNH
 
 | |
1GV8
 
 | | 18 kDa fragment of N-II domain of duck ovotransferrin | | Descriptor: | CARBONATE ION, FE (III) ION, GLYCINE, ... | | Authors: | Kuser, P, Hall, D.R, Haw, M.L, Neu, M, Lindley, P.F. | | Deposit date: | 2002-02-07 | | Release date: | 2002-02-12 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Mechanism of Iron Uptake by Transferrins: The X-Ray Structures of the 18 kDa Nii Domain Fragment of Duck Ovotransferrin and its Nitrilotriacetate Complex
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GVC
 
 | | 18kDa N-II domain fragment of duck ovotransferrin + NTA | | Descriptor: | CARBONATE ION, FE (III) ION, NITRILOTRIACETIC ACID, ... | | Authors: | Kuser, P, Hall, D.R, Haw, M.L, Neu, M, Lindley, P.F. | | Deposit date: | 2002-02-07 | | Release date: | 2002-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Mechanism of Iron Uptake by Transferrins: The X-Ray Structures of the 18 kDa Nii Domain Fragment of Duck Ovotransferrin and its Nitrilotriacetate Complex
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2YG8
 
 | | Structure of an unusual 3-Methyladenine DNA Glycosylase II (Alka) from Deinococcus radiodurans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DNA-3-methyladenine glycosidase II, ... | | Authors: | Moe, E, Hall, D.R, Leiros, I, Talstad, V, Timmins, J, McSweeney, S. | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-20 | | Last modified: | 2018-12-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
2CHV
 
 | | Replication Factor C ADPNP complex | | Descriptor: | REPLICATION FACTOR C SMALL SUBUNIT | | Authors: | Seybert, A, Singleton, M.R, Cook, N, Hall, D.R, Wigley, D.B. | | Deposit date: | 2006-03-16 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Communication between Subunits within an Archaeal Clamp-Loader Complex.
Embo J., 25, 2006
|
|
2YG9
 
 | | Structure of an unusual 3-Methyladenine DNA Glycosylase II (Alka) from Deinococcus radiodurans | | Descriptor: | CHLORIDE ION, DNA-3-methyladenine glycosidase II, putative, ... | | Authors: | Moe, E, Hall, D.R, Leiros, I, Talstad, V, Timmins, J, McSweeney, S. | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
2CHQ
 
 | | Replication Factor C ADPNP complex | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, REPLICATION FACTOR C SMALL SUBUNIT | | Authors: | Seybert, A, Singleton, M.R, Cook, N, Hall, D.R, Wigley, D.B. | | Deposit date: | 2006-03-16 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Communication between Subunits within an Archaeal Clamp-Loader Complex.
Embo J., 25, 2006
|
|
2CHG
 
 | | Replication Factor C domains 1 and 2 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, REPLICATION FACTOR C SMALL SUBUNIT | | Authors: | Seybert, A, Singleton, M.R, Cook, N, Hall, D.R, Wigley, D.B. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Communication between Subunits within an Archaeal Clamp-Loader Complex.
Embo J., 25, 2006
|
|
1W3S
 
 | | The crystal structure of RecO from Deinococcus radiodurans. | | Descriptor: | HYPOTHETICAL PROTEIN DR0819, ZINC ION | | Authors: | Leiros, I, Timmins, J, Hall, D.R, Leonard, G.A, McSweeney, S.M. | | Deposit date: | 2004-07-18 | | Release date: | 2005-02-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and DNA-Binding Analysis of Reco from Deinococcus Radiodurans
Embo J., 24, 2005
|
|
1QNG
 
 | |
2VLI
 
 | | Structure of Deinococcus radiodurans tunicamycin resistance protein | | Descriptor: | ANTIBIOTIC RESISTANCE PROTEIN, CADMIUM ION, CHLORIDE ION | | Authors: | Macedo, S, Kapp, U, Leiros, I, Hall, D.R, Mitchell, E. | | Deposit date: | 2008-01-15 | | Release date: | 2008-06-17 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Deinococcus Radiodurans Tunicamycin-Resistance Protein (Tmrd), a Phosphotransferase.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2UVP
 
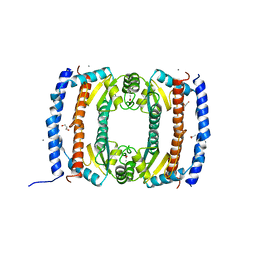 | | Crystal structure of HobA (HP1230)from Helicobacter pylori | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Terradot, L, Natrajan, G, Thompson, A.C, Hall, D.R. | | Deposit date: | 2007-03-13 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural similarity between the DnaA-binding proteins HobA (HP1230) from Helicobacter pylori and DiaA from Escherichia coli.
Mol. Microbiol., 65, 2007
|
|
4IT6
 
 | |
4UNF
 
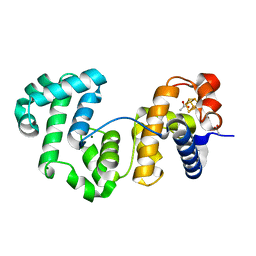 | | Crystal structure of Deinococcus radiodurans Endonuclease III-1 | | Descriptor: | ACETATE ION, ENDONUCLEASE III-1, IRON/SULFUR CLUSTER, ... | | Authors: | Sarre, A, Okvist, M, Klar, T, Hall, D, Smalas, A.O, McSweeney, S, Timmins, J, Moe, E. | | Deposit date: | 2014-05-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Characterization of Two Unusual Endonuclease III Enzymes from Deinococcus Radiodurans.
J.Struct.Biol., 191, 2015
|
|
8QZR
 
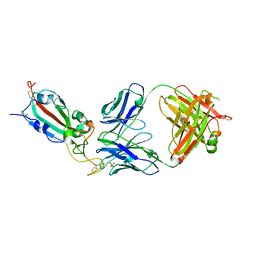 | | SARS-CoV-2 delta RBD complexed with BA.4/5-9 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-9 heavy chain, BA.4/5-9 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-10-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
8C3V
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.2-13 Fab and C1 nanobody | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, BA.2-13 heavy chain, BA.2-13 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-12-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
5WFL
 
 | | Kelch domain of human Keap1 in open unliganded conformation | | Descriptor: | Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Carolan, J.P, Lynch, A.J, Allen, K.N. | | Deposit date: | 2017-07-12 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Interaction Energetics and Druggability of the Protein-Protein Interaction between Kelch-like ECH-Associated Protein 1 (KEAP1) and Nuclear Factor Erythroid 2 Like 2 (Nrf2).
Biochemistry, 59, 2020
|
|
7QNY
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with COVOX-58 and COVOX-158 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 heavy chain, COVOX-158 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
