1A75
 
 | | WHITING PARVALBUMIN | | Descriptor: | CALCIUM ION, PARVALBUMIN | | Authors: | Declercq, J.P, Baneres, J.L, Rambaud, J, Parello, J. | | Deposit date: | 1998-03-19 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tertiary Structure of a Trp-Containing Parvalbumin from Whiting (Merlangius Merlangus). Description of the Hydrophobic Core
To be Published
|
|
2PVB
 
 | |
4PAL
 
 | | IONIC INTERACTIONS WITH PARVALBUMINS. CRYSTAL STRUCTURE DETERMINATION OF PIKE 4.10 PARVALBUMIN IN FOUR DIFFERENT IONIC ENVIRONMENTS | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PARVALBUMIN | | Authors: | Declercq, J.P, Tinant, B, Parello, J, Rambaud, J. | | Deposit date: | 1990-11-08 | | Release date: | 1992-01-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ionic interactions with parvalbumins. Crystal structure determination of pike 4.10 parvalbumin in four different ionic environments.
J.Mol.Biol., 220, 1991
|
|
2PAL
 
 | | IONIC INTERACTIONS WITH PARVALBUMINS. CRYSTAL STRUCTURE DETERMINATION OF PIKE 4.10 PARVALBUMIN IN FOUR DIFFERENT IONIC ENVIRONMENTS | | Descriptor: | MANGANESE (II) ION, PARVALBUMIN | | Authors: | Declercq, J.P, Tinant, B, Parello, J, Rambaud, J. | | Deposit date: | 1990-11-08 | | Release date: | 1992-01-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ionic interactions with parvalbumins. Crystal structure determination of pike 4.10 parvalbumin in four different ionic environments.
J.Mol.Biol., 220, 1991
|
|
3PAL
 
 | | IONIC INTERACTIONS WITH PARVALBUMINS. CRYSTAL STRUCTURE DETERMINATION OF PIKE 4.10 PARVALBUMIN IN FOUR DIFFERENT IONIC ENVIRONMENTS | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PARVALBUMIN | | Authors: | Declercq, J.P, Tinant, B, Parello, J, Rambaud, J. | | Deposit date: | 1990-11-08 | | Release date: | 1992-01-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ionic interactions with parvalbumins. Crystal structure determination of pike 4.10 parvalbumin in four different ionic environments.
J.Mol.Biol., 220, 1991
|
|
1PVB
 
 | | X-RAY STRUCTURE OF A NEW CRYSTAL FORM OF PIKE 4.10 PARVALBUMIN | | Descriptor: | AMMONIUM ION, CALCIUM ION, PARVALBUMIN | | Authors: | Declercq, J.P, Tinant, B, Parello, J. | | Deposit date: | 1995-01-05 | | Release date: | 1995-02-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray structure of a new crystal form of pike 4.10 beta parvalbumin.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1HD2
 
 | | Human peroxiredoxin 5 | | Descriptor: | BENZOIC ACID, BROMIDE ION, PEROXIREDOXIN 5 RESIDUES 54-214 | | Authors: | Declercq, J.P, Evrard, C. | | Deposit date: | 2000-11-06 | | Release date: | 2001-08-28 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Human Peroxiredoxin 5, a Novel Type of Mammalian Peroxiredoxin at 1.5 A Resolution.
J.Mol.Biol., 311, 2001
|
|
1PAL
 
 | | IONIC INTERACTIONS WITH PARVALBUMINS. CRYSTAL STRUCTURE DETERMINATION OF PIKE 4.10 PARVALBUMIN IN FOUR DIFFERENT IONIC ENVIRONMENTS | | Descriptor: | AMMONIUM ION, CALCIUM ION, PARVALBUMIN | | Authors: | Declercq, J.P, Tinant, B, Parello, J, Rambaud, J. | | Deposit date: | 1990-11-08 | | Release date: | 1992-01-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Ionic interactions with parvalbumins. Crystal structure determination of pike 4.10 parvalbumin in four different ionic environments.
J.Mol.Biol., 220, 1991
|
|
1H4O
 
 | | Monoclinic form of human peroxiredoxin 5 | | Descriptor: | BENZOIC ACID, PEROXIREDOXIN 5 | | Authors: | Declercq, J.P, Evrard, C. | | Deposit date: | 2001-05-11 | | Release date: | 2001-10-23 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Human Peroxiredoxin 5, a Novel Type of Mammalian Peroxiredoxin at 1.5 A Resolution
J.Mol.Biol., 311, 2001
|
|
2WLR
 
 | |
2WLX
 
 | |
1URM
 
 | | HUMAN PEROXIREDOXIN 5, C47S MUTANT | | Descriptor: | BENZOIC ACID, CHLORIDE ION, PEROXIREDOXIN 5 | | Authors: | Declercq, J.P, Evrard, C. | | Deposit date: | 2003-10-31 | | Release date: | 2004-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the C47S Mutant of Human Peroxiredoxin 5
J. Chem. Cryst., 34, 2004
|
|
1PVA
 
 | | COMPARISON BETWEEN THE CRYSTAL AND THE SOLUTION STRUCTURES OF THE EF HAND PARVALBUMIN (ALPHA COMPONENT FROM PIKE MUSCLE) | | Descriptor: | CALCIUM ION, PARVALBUMIN | | Authors: | Declercq, J.P, Tinant, B, Roquet, F, Rambaud, J, Parello, J. | | Deposit date: | 1995-01-16 | | Release date: | 1995-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Comparison between the Crystal and the Solution Structures of the EF Hand Parvalbumin (Alpha Component from Pike Muscle)
To be Published
|
|
2Y1B
 
 | | Crystal structure of the E. coli outer membrane lipoprotein RcsF | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, PUTATIVE OUTER MEMBRANE PROTEIN, SIGNAL, ... | | Authors: | Declercq, J.P, Leverrier, P, Boujtat, A, Collet, J.F. | | Deposit date: | 2010-12-07 | | Release date: | 2011-03-16 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Outer Membrane Protein Rcsf, a New Substrate for the Periplasmic Protein- Disulfide Isomerase Dsbc.
J.Biol.Chem., 286, 2011
|
|
1AM7
 
 | | Lysozyme from bacteriophage lambda | | Descriptor: | ISOPROPYL ALCOHOL, LYSOZYME | | Authors: | Evrard, C, Fastrez, J, Declercq, J.P. | | Deposit date: | 1997-06-24 | | Release date: | 1997-12-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the lysozyme from bacteriophage lambda and its relationship with V and C-type lysozymes.
J.Mol.Biol., 276, 1998
|
|
1UVZ
 
 | |
1W4V
 
 | |
1W89
 
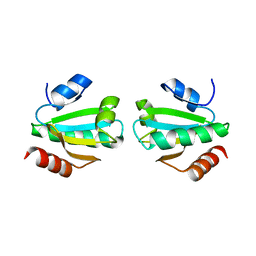 | |
2J8Y
 
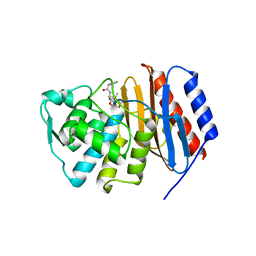 | | Structure of PBP-A acyl-enzyme complex with penicillin-G | | Descriptor: | OPEN FORM - PENICILLIN G, TLL2115 PROTEIN | | Authors: | Evrard, C, Declercq, J.P. | | Deposit date: | 2006-10-31 | | Release date: | 2007-11-27 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Pbp-A from Thermosynechococcus Elongatus, a Penicillin-Binding Protein Closely Related to Class a Beta-Lactamases.
J.Mol.Biol., 386, 2009
|
|
2J9O
 
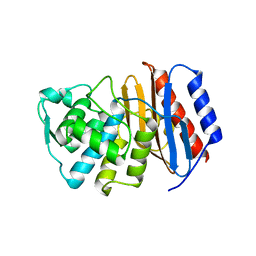 | | Structure of PBP-A, L158E mutant | | Descriptor: | GLYCEROL, TLL2115 PROTEIN | | Authors: | Evrard, C, Declercq, J.P. | | Deposit date: | 2006-11-14 | | Release date: | 2007-12-11 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Pbp-A from Thermosynechococcus Elongatus, a Penicillin-Binding Protein Closely Related to Class a Beta-Lactamases.
J.Mol.Biol., 386, 2009
|
|
2IU2
 
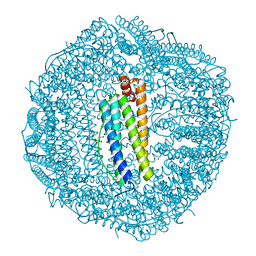 | | Recombinant human H ferritin, K86Q, E27D and E107D mutant, soaked with Zn ions | | Descriptor: | FERRITIN HEAVY CHAIN, GLYCEROL, ZINC ION | | Authors: | Toussaint, L, Crichton, R.R, Declercq, J.P. | | Deposit date: | 2006-05-26 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-Resolution X-Ray Structures of Human Apoferritin H-Chain Mutants Correlated with Their Activity and Metal-Binding Sites.
J.Mol.Biol., 365, 2007
|
|
2J7V
 
 | | Structure of PBP-A | | Descriptor: | TLL2115 PROTEIN | | Authors: | Evrard, C, Declercq, J.P. | | Deposit date: | 2006-10-17 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Pbp-A from Thermosynechococcus Elongatus, a Penicillin-Binding Protein Closely Related to Class a Beta-Lactamases.
J.Mol.Biol., 386, 2009
|
|
2CN7
 
 | | Recombinant human H ferritin, K86Q, E27D and E107D mutant | | Descriptor: | CALCIUM ION, FERRITIN HEAVY CHAIN, GLYCEROL | | Authors: | Toussaint, L, Crichton, R.R, Declercq, J.P. | | Deposit date: | 2006-05-18 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-Resolution X-Ray Structures of Human Apoferritin H-Chain Mutants Correlated with Their Activity and Metal-Binding Sites.
J.Mol.Biol., 365, 2007
|
|
2CHI
 
 | | Recombinant human H ferritin, K86Q and E27D mutant | | Descriptor: | CALCIUM ION, FERRITIN HEAVY CHAIN, GLYCEROL, ... | | Authors: | Toussaint, L, Crichton, R.R, Declercq, J.P. | | Deposit date: | 2006-03-15 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-Resolution X-Ray Structures of Human Apoferritin H-Chain Mutants Correlated with Their Activity and Metal-Binding Sites.
J.Mol.Biol., 365, 2007
|
|
2CEI
 
 | | Recombinant human H ferritin, K86Q mutant, soaked with Zn | | Descriptor: | FERRITIN HEAVY CHAIN, ZINC ION | | Authors: | Toussaint, L, Crichton, R.R, Declercq, J.P. | | Deposit date: | 2006-02-07 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-Resolution X-Ray Structures of Human Apoferritin H-Chain Mutants Correlated with Their Activity and Metal-Binding Sites.
J.Mol.Biol., 365, 2007
|
|
