2GBH
 
 | | NMR structure of stem region of helix-35 of 23S E.coli ribosomal RNA (residues 736-760) | | Descriptor: | 5'-R(*(GMP)P*GP*GP*CP*UP*AP*AP*UP*GP*(PSU)P*UP*GP*AP*AP*AP*AP*AP*UP*UP*AP*GP*CP*CP*C)-3' | | Authors: | Bax, A, Boisbouvier, J, Bryce, D, Grishaev, A, Jaroniec, C, Miclet, E, Nikonovicz, E, O'Neil-Cabello, E, Ying, J. | | Deposit date: | 2006-03-10 | | Release date: | 2006-04-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Measurement of five dipolar couplings from a single 3D NMR multiplet applied to the study of RNA dynamics.
J.Am.Chem.Soc., 126, 2004
|
|
6OBI
 
 | | Remarkable rigidity of the single alpha-helical domain of myosin-VI revealed by NMR spectroscopy | | Descriptor: | Myosin-VI | | Authors: | Barnes, A, Shen, Y, Ying, J, Takagi, Y, Torchia, D.A, Sellers, J, Bax, A. | | Deposit date: | 2019-03-20 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Remarkable Rigidity of the Single alpha-Helical Domain of Myosin-VI As Revealed by NMR Spectroscopy.
J.Am.Chem.Soc., 141, 2019
|
|
4D1Q
 
 | | Hermes transposase bound to its terminal inverted repeat | | Descriptor: | SODIUM ION, TERMINAL INVERTED REPEAT, TRANSPOSASE | | Authors: | Hickman, A.B, Ewis, H, Li, X, Knapp, J, Laver, T, Doss, A.L, Tolun, G, Steven, A, Grishaev, A, Bax, A, Atkinson, P, Craig, N.L, Dyda, F. | | Deposit date: | 2014-05-04 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Hat Transposon End Recognition by Hermes, an Octameric DNA Transposase from Musca Domestica.
Cell(Cambridge,Mass.), 158, 2014
|
|
2A5M
 
 | |
1HKS
 
 | | SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF DROSOPHILA HEAT SHOCK TRANSCRIPTION FACTOR | | Descriptor: | HEAT-SHOCK TRANSCRIPTION FACTOR | | Authors: | Vuister, G.W, Kim, S.-J, Orosz, A, Marquardt, J.L, Wu, C, Bax, A. | | Deposit date: | 1994-07-18 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of Drosophila heat shock transcription factor.
Nat.Struct.Biol., 1, 1994
|
|
1HKT
 
 | | SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF DROSOPHILA HEAT SHOCK TRANSCRIPTION FACTOR | | Descriptor: | HEAT-SHOCK TRANSCRIPTION FACTOR | | Authors: | Vuister, G.W, Kim, S.-J, Orosz, A, Marquardt, J.L, Wu, C, Bax, A. | | Deposit date: | 1994-07-18 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of Drosophila heat shock transcription factor.
Nat.Struct.Biol., 1, 1994
|
|
7R95
 
 | |
6B3U
 
 | | Solution Structure of HIV-1 GP41 Transmembrane Domain in Bicelles | | Descriptor: | HIV-1 GP41 Transmembrane Domain | | Authors: | Chiliveri, S.C, Louis, J.M, Ghirlando, R, Baber, J.L, Bax, A. | | Deposit date: | 2017-09-24 | | Release date: | 2018-01-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Tilted, Uninterrupted, Monomeric HIV-1 gp41 Transmembrane Helix from Residual Dipolar Couplings.
J. Am. Chem. Soc., 140, 2018
|
|
1ZWO
 
 | | NMR structure of murine gamma-S crystallin | | Descriptor: | Gamma crystallin S | | Authors: | Wu, Z, Delaglio, F, Wyatt, K, Wistow, G, Bax, A. | | Deposit date: | 2005-06-04 | | Release date: | 2005-07-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of (gamma)S-crystallin by molecular fragment replacement NMR.
Protein Sci., 14, 2005
|
|
1ZWM
 
 | | NMR structure of murine gamma-S crystallin | | Descriptor: | Gamma crystallin S | | Authors: | Wu, Z, Delaglio, F, Wyatt, K, Wistow, G, Bax, A. | | Deposit date: | 2005-06-03 | | Release date: | 2005-07-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of (gamma)S-crystallin by molecular fragment replacement NMR.
Protein Sci., 14, 2005
|
|
1DUF
 
 | | THE NMR STRUCTURE OF DNA DODECAMER DETERMINED IN AQUEOUS DILUTE LIQUID CRYSTALLINE PHASE | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Tjandra, N, Tate, S, Ono, A, Kainosho, M, Bax, A. | | Deposit date: | 2000-01-17 | | Release date: | 2000-07-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of a DNA Dodecamer in an Aqueous Dilute Liquid Crystalline Phase
J.Am.Chem.Soc., 122, 2000
|
|
1F71
 
 | |
1F70
 
 | |
2BBM
 
 | | SOLUTION STRUCTURE OF A CALMODULIN-TARGET PEPTIDE COMPLEX BY MULTIDIMENSIONAL NMR | | Descriptor: | CALCIUM ION, CALMODULIN, MYOSIN LIGHT CHAIN KINASE | | Authors: | Clore, G.M, Bax, A, Ikura, M, Gronenborn, A.M. | | Deposit date: | 1992-07-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a calmodulin-target peptide complex by multidimensional NMR.
Science, 256, 1992
|
|
2BBN
 
 | | SOLUTION STRUCTURE OF A CALMODULIN-TARGET PEPTIDE COMPLEX BY MULTIDIMENSIONAL NMR | | Descriptor: | CALCIUM ION, CALMODULIN, MYOSIN LIGHT CHAIN KINASE | | Authors: | Clore, G.M, Bax, A, Ikura, M, Gronenborn, A.M. | | Deposit date: | 1992-07-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a calmodulin-target peptide complex by multidimensional NMR.
Science, 256, 1992
|
|
2ARI
 
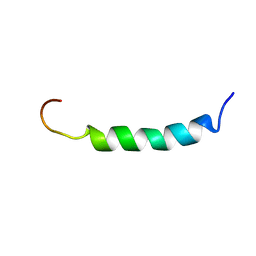 | | Solution structure of micelle-bound fusion domain of HIV-1 gp41 | | Descriptor: | Envelope polyprotein GP160 | | Authors: | Jaroniec, C.P, Kaufman, J.D, Stahl, S.J, Viard, M, Blumenthal, R, Wingfield, P.T, Bax, A. | | Deposit date: | 2005-08-19 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of Micelle-Associated Human Immunodeficiency Virus gp41 Fusion Domain.
Biochemistry, 44, 2005
|
|
1XQ8
 
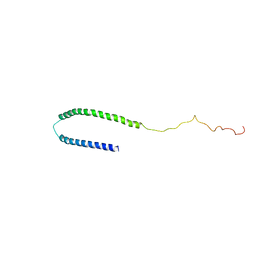 | |
1J7O
 
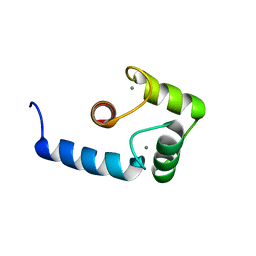 | |
1J7P
 
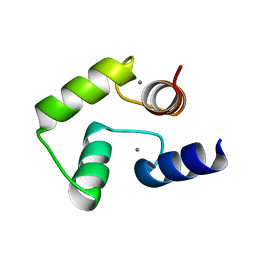 | |
1LVZ
 
 | | METARHODOPSIN II BOUND STRUCTURE OF C-TERMINAL PEPTIDE OF ALPHA-SUBUNIT OF TRANSDUCIN | | Descriptor: | Guanine nucleotide-binding protein G(T), alpha-1 subunit | | Authors: | Koenig, B.W, Kontaxis, G, Mitchell, D.C, Louis, J.M, Litman, B.J, Bax, A. | | Deposit date: | 2002-05-30 | | Release date: | 2002-09-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and orientation of a G protein fragment in the receptor bound state from residual dipolar couplings.
J.Mol.Biol., 322, 2002
|
|
1P7E
 
 | |
1P7F
 
 | |
1D3Z
 
 | | UBIQUITIN NMR STRUCTURE | | Descriptor: | PROTEIN (UBIQUITIN) | | Authors: | Cornilescu, G, Marquardt, J.L, Ottiger, M, Bax, A. | | Deposit date: | 1999-10-01 | | Release date: | 1999-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Validation of Protein Structure from Anisotropic Carbonyl Chemical Shifts in a Dilute Liquid Crystalline Phase
J.Am.Chem.Soc., 120, 1998
|
|
1CFD
 
 | | CALCIUM-FREE CALMODULIN | | Descriptor: | CALMODULIN | | Authors: | Kuboniwa, H, Tjandra, N, Grzesiek, S, Ren, H, Klee, C.B, Bax, A. | | Deposit date: | 1995-10-18 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calcium-free calmodulin.
Nat.Struct.Biol., 2, 1995
|
|
1CFC
 
 | | CALCIUM-FREE CALMODULIN | | Descriptor: | CALMODULIN | | Authors: | Kuboniwa, H, Tjandra, N, Grzesiek, S, Ren, H, Klee, C.B, Bax, A. | | Deposit date: | 1995-08-02 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calcium-free calmodulin.
Nat.Struct.Biol., 2, 1995
|
|
