1F2D
 
 | | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE DEAMINASE | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE DEAMINASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Yao, M, Ose, T, Sugimoto, H, Horiuchi, A, Nakagawa, A, Yokoi, D, Murakami, T, Honma, M, Wakatsuki, S, Tanaka, I. | | Deposit date: | 2000-05-24 | | Release date: | 2000-12-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 1-aminocyclopropane-1-carboxylate deaminase from Hansenula saturnus.
J.Biol.Chem., 275, 2000
|
|
1IZ6
 
 | | Crystal Structure of Translation Initiation Factor 5A from Pyrococcus Horikoshii | | Descriptor: | Initiation Factor 5A | | Authors: | Yao, M, Ohsawa, A, Kikukawa, S, Tanaka, I, Kimura, M. | | Deposit date: | 2002-09-25 | | Release date: | 2003-01-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Hyperthermophilic Archaeal Initiation Factor 5A: A Homologue of Eukaryotic Initiation Factor 5A (eIF-5A)
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1WLS
 
 | | Crystal structure of L-asparaginase I homologue protein from Pyrococcus horikoshii | | Descriptor: | L-asparaginase | | Authors: | Yao, M, Morita, H, Yasutake, Y, Tanaka, I. | | Deposit date: | 2004-06-29 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of the type I L-asparaginase from the hyperthermophilic archaeon Pyrococcus horikoshii at 2.16 angstroms resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2ZQ0
 
 | | Crystal structure of SusB complexed with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase (Alpha-glucosidase SusB), CALCIUM ION | | Authors: | Yao, M, Tanaka, I, Kitamura, M. | | Deposit date: | 2008-07-31 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides thetaiotaomicron.
J.Biol.Chem., 283, 2008
|
|
1V7L
 
 | |
1VGJ
 
 | | Crystal structure of 2'-5' RNA ligase from Pyrococcus horikoshii | | Descriptor: | Hypothetical protein PH0099 | | Authors: | Yao, M, Morita, H, Okada, A, Tanaka, I. | | Deposit date: | 2004-04-27 | | Release date: | 2005-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The structure of Pyrococcus horikoshii 2'-5' RNA ligase at 1.94 A resolution reveals a possible open form with a wider active-site cleft
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
1EL1
 
 | |
1OCO
 
 | |
4P1Y
 
 | | Crystal structure of staphylococcal gamma-hemolysin prepore | | Descriptor: | Gamma-hemolysin component A, Gamma-hemolysin component B | | Authors: | Yamashita, D, Tanaka, Y, Tanaka, I, Yao, M. | | Deposit date: | 2014-02-28 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.992 Å) | | Cite: | Molecular basis of transmembrane beta-barrel formation of staphylococcal pore-forming toxins.
Nat Commun, 5, 2014
|
|
4P1X
 
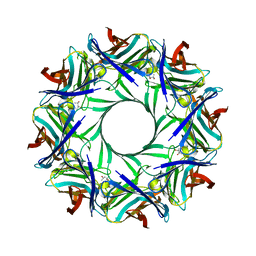 | | Crystal structure of staphylococcal LUK prepore | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Gamma-hemolysin component B, Gamma-hemolysin component C | | Authors: | Yamashita, D, Tanaka, Y, Tanaka, I, Yao, M. | | Deposit date: | 2014-02-28 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of transmembrane beta-barrel formation of staphylococcal pore-forming toxins.
Nat Commun, 5, 2014
|
|
4WLC
 
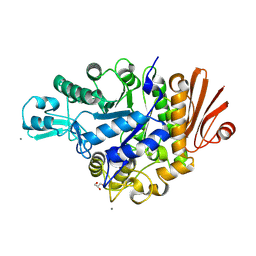 | | Structure of dextran glucosidase with glucose | | Descriptor: | CALCIUM ION, GLYCEROL, Glucan 1,6-alpha-glucosidase, ... | | Authors: | Kobayashi, M, Kato, K, Yao, M. | | Deposit date: | 2014-10-07 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structural insights into the catalytic reaction that is involved in the reorientation of Trp238 at the substrate-binding site in GH13 dextran glucosidase
Febs Lett., 589, 2015
|
|
4P24
 
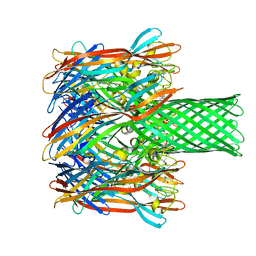 | | pore forming toxin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha-hemolysin | | Authors: | Sugawara, T, Yamashita, D, Tanaka, Y, Tanaka, I, Yao, M. | | Deposit date: | 2014-03-01 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for pore-forming mechanism of staphylococcal alpha-hemolysin.
Toxicon, 108, 2015
|
|
4V60
 
 | | The structure of rat liver vault at 3.5 angstrom resolution | | Descriptor: | Major vault protein | | Authors: | Kato, K, Zhou, Y, Tanaka, H, Yao, M, Yamashita, E, Yoshimura, M, Tsukihara, T. | | Deposit date: | 2008-10-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of rat liver vault at 3.5 angstrom resolution
Science, 323, 2009
|
|
5IP3
 
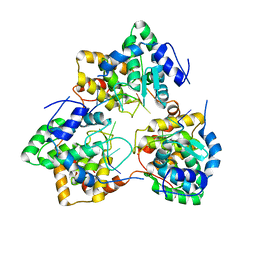 | | Tomato spotted wilt tospovirus nucleocapsid protein-ssDNA complex | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*T)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Komoda, K, Narita, M, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Asymmetric Trimeric Ring Structure of the Nucleocapsid Protein of Tospovirus.
J. Virol., 91, 2017
|
|
5IP2
 
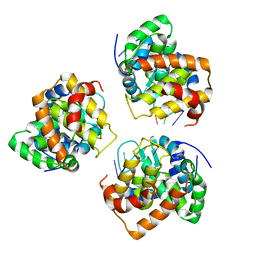 | | Tomato spotted wilt tospovirus nucleocapsid protein-ssRNA complex | | Descriptor: | Nucleoprotein, RNA (5'-D(P*UP*UP*U)-3'), RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Komoda, K, Narita, M, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Asymmetric Trimeric Ring Structure of the Nucleocapsid Protein of Tospovirus.
J. Virol., 91, 2017
|
|
4WJ3
 
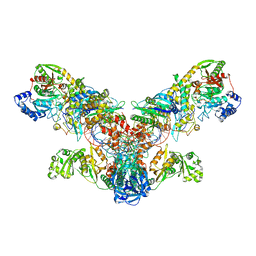 | | Crystal structure of the asparagine transamidosome from Pseudomonas aeruginosa | | Descriptor: | 76mer-tRNA, Aspartate--tRNA(Asp/Asn) ligase, Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, ... | | Authors: | Suzuki, T, Nakamura, A, Kato, K, Tanaka, I, Yao, M. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-31 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (3.705 Å) | | Cite: | Structure of the Pseudomonas aeruginosa transamidosome reveals unique aspects of bacterial tRNA-dependent asparagine biosynthesis
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8IN4
 
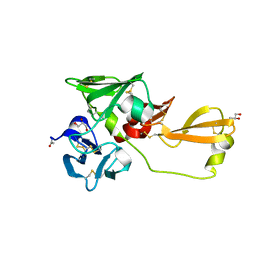 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | Descriptor: | 25 kDa polyphenol-binding protein, ACETYL GROUP, GLYCEROL | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN1
 
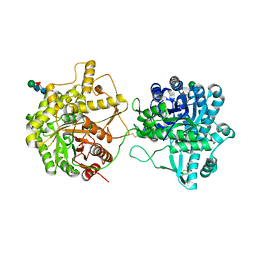 | | beta-glucosidase protein from Aplysia kurodai | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-Glucosidase, alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN3
 
 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | Descriptor: | 25 kDa polyphenol-binding protein, GLYCEROL | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN6
 
 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai complex with tannic acid | | Descriptor: | 25 kDa polyphenol-binding protein, BETA-1,2,3,4,6-PENTA-O-GALLOYL-D-GLUCOPYRANOSE | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
3VO9
 
 | | Staphylococcus aureus FtsZ apo-form (SeMet) | | Descriptor: | Cell division protein FtsZ | | Authors: | Matsui, T, Yamane, J, Mogi, N, Yao, M, Tanaka, I. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-29 | | Last modified: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8YIE
 
 | | Crystal structure of GH13_30 alpha-glucosidase CmmB in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase | | Authors: | Saburi, W, Tagami, T, Yu, J, Ose, T, Yao, M, Mori, H. | | Deposit date: | 2024-02-29 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular mechanism for the substrate specificity of Arthrobacter globiformis M6 alpha-glucosidase CmmB, belonging to glycoside hydrolase family 13 subfamily 30
Food Biosci, 61, 2024
|
|
8YIF
 
 | | Crystal structure of GH13_30 alpha-glucosidase CmmB in complex with acarviosin | | Descriptor: | Acarviosin, Alpha-glucosidase | | Authors: | Saburi, W, Tagami, T, Yu, J, Ose, T, Yao, M, Mori, H. | | Deposit date: | 2024-02-29 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular mechanism for the substrate specificity of Arthrobacter globiformis M6 alpha-glucosidase CmmB, belonging to glycoside hydrolase family 13 subfamily 30
Food Biosci, 61, 2024
|
|
3VOA
 
 | | Staphylococcus aureus FtsZ 12-316 GDP-form | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yamane, J, Matsui, T, Mogi, N, Yao, M, Tanaka, I. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VZR
 
 | |
