2F07
 
 | |
3L9F
 
 | |
3LBE
 
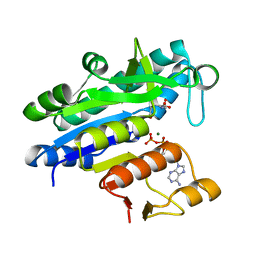
 | | The Crystal Structure of smu.793 from Streptococcus mutans UA159 bound to acetyl CoA | | Descriptor: | CHLORIDE ION, COENZYME A, Putative uncharacterized protein smu.793 | | Authors: | Su, X.-D, Hou, Q.M, Fan, X.X, Nan, J, Liu, X. | | Deposit date: | 2010-01-08 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of smu.793 from Streptococcus mutans UA159 bound to acetyl CoA
TO BE PUBLISHED
|
|
3L86
 
 | |
3L9D
 
 | |
3L7Y
 
 | |
3LD2
 
 | |
3L7X
 
 | |
3L87
 
 | |
3L9C
 
 | |
3LA8
 
 | |
3LBB
 
 | | The Crystal Structure of smu.793 from Streptococcus mutans UA159 | | Descriptor: | CHLORIDE ION, Putative uncharacterized protein smu.793, SULFATE ION | | Authors: | Su, X.-D, Hou, Q.M, Fan, X.X, Nan, J, Liu, X. | | Deposit date: | 2010-01-08 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of smu.793 from Streptococcus mutans UA159
TO BE PUBLISHED
|
|
3L7W
 
 | |
3L9T
 
 | |
3LBA
 
 | |
3LEH
 
 | |
1PHR
 
 | | THE CRYSTAL STRUCTURE OF A LOW MOLECULAR PHOSPHOTYROSINE PROTEIN PHOSPHATASE | | Descriptor: | LOW MOLECULAR WEIGHT PHOSPHOTYROSINE PROTEIN PHOSPHATASE, SULFATE ION | | Authors: | Su, X.-D, Taddei, N, Stefani, M, Ramponi, G, Nordlund, P. | | Deposit date: | 1994-07-05 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a low-molecular-weight phosphotyrosine protein phosphatase.
Nature, 370, 1994
|
|
3S2S
 
 | |
3S70
 
 | | Crystal structure of active caspase-6 bound with Ac-VEID-CHO solved by As-SAD | | Descriptor: | ACETATE ION, CACODYLATE ION, Caspase-6, ... | | Authors: | Su, X.-D, Liu, X, Wang, X.-J. | | Deposit date: | 2011-05-26 | | Release date: | 2012-04-11 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.625 Å) | | Cite: | Get phases from arsenic anomalous scattering: de novo SAD phasing of two protein structures crystallized in cacodylate buffer
Plos One, 6, 2011
|
|
3NR2
 
 | | Crystal structure of Caspase-6 zymogen | | Descriptor: | Caspase-6 | | Authors: | Su, X.-D, Wang, X.-J, Liu, X, Mi, W, Wang, K.-T. | | Deposit date: | 2010-06-30 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation
Embo Rep., 11, 2010
|
|
3L7V
 
 | |
1HXJ
 
 | |
4F8Y
 
 | |
1LVA
 
 | |
2BAZ
 
 | |
