7X3K
 
 | |
4ANI
 
 | | Structural basis for the intermolecular communication between DnaK and GrpE in the DnaK chaperone system from Geobacillus kaustophilus HTA426 | | Descriptor: | CHAPERONE PROTEIN DNAK, PROTEIN GRPE | | Authors: | Wu, C.-C, Naveen, V, Chien, C.-H, Chang, Y.-W, Hsiao, C.-D. | | Deposit date: | 2012-03-19 | | Release date: | 2012-05-23 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (4.094 Å) | | Cite: | Crystal Structure of Dnak Protein Complexed with Nucleotide Exchange Factor Grpe in Dnak Chaperone System: Insight Into Intermolecular Communication.
J.Biol.Chem., 287, 2012
|
|
7Z3N
 
 | | Cryo-EM structure of the ribosome-associated RAC complex on the 80S ribosome - RAC-1 conformation | | Descriptor: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kisonaite, M, Wild, K, Sinning, I. | | Deposit date: | 2022-03-02 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural inventory of cotranslational protein folding by the eukaryotic RAC complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z3O
 
 | | Cryo-EM structure of the ribosome-associated RAC complex on the 80S ribosome - RAC-2 conformation | | Descriptor: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kisonaite, M, Wild, K, Sinning, I. | | Deposit date: | 2022-03-02 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural inventory of cotranslational protein folding by the eukaryotic RAC complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1XQS
 
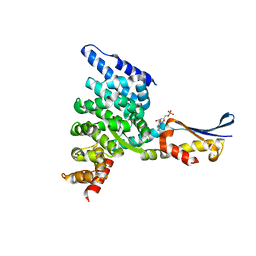 | | Crystal structure of the HspBP1 core domain complexed with the fragment of Hsp70 ATPase domain | | Descriptor: | ADENOSINE MONOPHOSPHATE, HSPBP1 protein, Heat shock 70 kDa protein 1 | | Authors: | Shomura, Y, Dragovic, Z, Chang, H.C, Tzvetkov, N, Young, J.C, Brodsky, J.L, Guerriero, V, Hartl, F.U, Bracher, A. | | Deposit date: | 2004-10-13 | | Release date: | 2005-03-01 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Regulation of Hsp70 Function by HspBP1; Structural Analysis Reveals an Alternate Mechanism for Hsp70 Nucleotide Exchange
Mol.Cell, 17, 2005
|
|
4E81
 
 | |
8GB3
 
 | |
8OO0
 
 | | Chaetomium thermophilum Methionine Aminopeptidase 2 autoproteolysis product at the 80S ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S0, ... | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
6H9U
 
 | | Crystal structure of the BiP NBD and MANF SAP complex | | Descriptor: | D-MALATE, Endoplasmic reticulum chaperone BiP, Mesencephalic astrocyte-derived neurotrophic factor, ... | | Authors: | Yan, Y, Ron, D. | | Deposit date: | 2018-08-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | MANF antagonizes nucleotide exchange by the endoplasmic reticulum chaperone BiP.
Nat Commun, 10, 2019
|
|
3QML
 
 | | The structural analysis of Sil1-Bip complex reveals the mechanism for Sil1 to function as a novel nucleotide exchange factor | | Descriptor: | 78 kDa glucose-regulated protein homolog, MAGNESIUM ION, Nucleotide exchange factor SIL1, ... | | Authors: | Yan, M, Li, J.Z, Sha, B.D. | | Deposit date: | 2011-02-04 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of the Sil1-Bip complex reveals the mechanism for Sil1 to function as a nucleotide-exchange factor.
Biochem.J., 438, 2011
|
|
1DKG
 
 | |
6HA7
 
 | | Crystal structure of the BiP NBD and MANF complex | | Descriptor: | 1,2-ETHANEDIOL, Endoplasmic reticulum chaperone BiP, Mesencephalic astrocyte-derived neurotrophic factor | | Authors: | Yan, Y, Ron, D. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | MANF antagonizes nucleotide exchange by the endoplasmic reticulum chaperone BiP.
Nat Commun, 10, 2019
|
|
3A8Y
 
 | | Crystal structure of the complex between the BAG5 BD5 and Hsp70 NBD | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG family molecular chaperone regulator 5, Heat shock 70 kDa protein 1 | | Authors: | Arakawa, A, Handa, N, Ohsawa, N, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-13 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange
Structure, 18, 2010
|
|
1HX1
 
 | | CRYSTAL STRUCTURE OF A BAG DOMAIN IN COMPLEX WITH THE HSC70 ATPASE DOMAIN | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG family molecular chaperone regulator 1, Heat shock 70 kDa protein 8 | | Authors: | Sondermann, H, Scheufler, C, Moarefi, I. | | Deposit date: | 2001-01-11 | | Release date: | 2001-03-07 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors.
Science, 291, 2001
|
|
3LDQ
 
 | | Crystal structure of HSC70/BAG1 in complex with small molecule inhibitor | | Descriptor: | 8-[(quinolin-2-ylmethyl)amino]adenosine, BAG family molecular chaperone regulator 1, Heat shock cognate 71 kDa protein | | Authors: | Dokurno, P, Surgenor, A.E, Shaw, T, Macias, A.T, Massey, A.J, Williamson, D.S. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adenosine-Derived Inhibitors of 78 kDa Glucose Regulated Protein (Grp78) ATPase: Insights into Isoform Selectivity.
J.Med.Chem., 54, 2011
|
|
3M3Z
 
 | | Crystal structure of HSC70/BAG1 in complex with small molecule inhibitor | | Descriptor: | 5'-O-(2-amino-2-oxoethyl)-8-(methylamino)adenosine, BAG family molecular chaperone regulator 1, Heat shock cognate 71 kDa protein | | Authors: | Dokurno, P, Surgenor, A.E, Shaw, T, Macias, A.T, Massey, A.J, Williamson, D.S. | | Deposit date: | 2010-03-10 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Adenosine-Derived Inhibitors of 78 kDa Glucose Regulated Protein (Grp78) ATPase: Insights into Isoform Selectivity.
J.Med.Chem., 54, 2011
|
|
6ZMD
 
 | | Crystal structure of HYPE covalently tethered to BiP bound to AMP-PNP | | Descriptor: | Endoplasmic reticulum chaperone BiP, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Fauser, J, Gulen, B, Pett, C, Hedberg, C, Itzen, A, Pogenberg, V. | | Deposit date: | 2020-07-02 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Specificity of AMPylation of the human chaperone BiP is mediated by TPR motifs of FICD.
Nat Commun, 12, 2021
|
|
5NRO
 
 | | Structure of full-length DnaK with bound J-domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaJ, Chaperone protein DnaK, ... | | Authors: | Kopp, J, Kityk, R, Mayer, M.P. | | Deposit date: | 2017-04-25 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Molecular Mechanism of J-Domain-Triggered ATP Hydrolysis by Hsp70 Chaperones.
Mol. Cell, 69, 2018
|
|
7B7Z
 
 | | DeAMPylation complex of monomeric FICD and AMPylated BiP (state 1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Endoplasmic reticulum chaperone BiP, ... | | Authors: | Perera, L.A, Ron, D. | | Deposit date: | 2020-12-12 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of a deAMPylation complex rationalise the switch between antagonistic catalytic activities of FICD.
Nat Commun, 12, 2021
|
|
7B80
 
 | | DeAMPylation complex of monomeric FICD and AMPylated BiP (state 2) | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Perera, L.A, Ron, D. | | Deposit date: | 2020-12-12 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structures of a deAMPylation complex rationalise the switch between antagonistic catalytic activities of FICD.
Nat Commun, 12, 2021
|
|
4F00
 
 | |
4EZU
 
 | |
7JN9
 
 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide QEHTGSQLRIAAYGP | | Descriptor: | Alkaline phosphatase peptide, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KW7
 
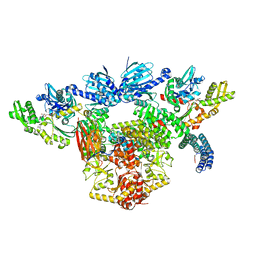 | | Atomic cryoEM structure of Hsp90-Hsp70-Hop-GR | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glucocorticoid receptor, Heat shock 70 kDa protein 1A, ... | | Authors: | Wang, R.Y, Noddings, C.M, Kirschke, E, Myasnikov, A, Johnson, J.L, Agard, D.A. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-08 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure of Hsp90-Hsp70-Hop-GR reveals the Hsp90 client-loading mechanism.
Nature, 601, 2022
|
|
3FZK
 
 | | Crystal Structures of Hsc70/Bag1 in Complex with Small Molecule Inhibitors | | Descriptor: | (2R,3R,4S,5R)-2-[6-amino-8-[(3,4-dichlorophenyl)methylamino]purin-9-yl]-5-(hydroxymethyl)oxolane-3,4-diol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG family molecular chaperone regulator 1, ... | | Authors: | Dokurno, P, Williamson, D.S, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel adenosine-derived inhibitors of 70 kDa heat shock protein, discovered through structure-based design
J.Med.Chem., 52, 2009
|
|
