3ICZ
 
 | | Trypanosoma cruzi farnesyl diphosphate synthase homodimer in complex with 1-(2,2-Bis-phosphono-ethyl)-3-butyl-pyridinium and isopentenyl pyrophosphate | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, 3-[(1E)-but-1-en-1-yl]-1-(2,2-diphosphonoethyl)pyridinium, Farnesyl pyrophosphate synthase, ... | | Authors: | Amzel, L.M, Huang, C.H, Gabelli, S.B, Oldfield, E. | | Deposit date: | 2009-07-19 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Binding of nitrogen-containing bisphosphonates (N-BPs) to the Trypanosoma cruzi farnesyl diphosphate synthase homodimer.
Proteins, 78, 2010
|
|
3IJO
 
 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, althiazide | | Descriptor: | (3S)-6-chloro-3-[(prop-2-en-1-ylsulfanyl)methyl]-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ptak, C.P, Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3IR5
 
 | | Crystal structure of NarGHI mutant NarG-H49C | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2009-08-21 | | Release date: | 2010-01-05 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein crystallography reveals a role for the FS0 cluster of Escherichia coli nitrate reductase A (NarGHI) in enzyme maturation.
J.Biol.Chem., 285, 2010
|
|
3IR7
 
 | | Crystal structure of NarGHI mutant NarG-R94S | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2009-08-21 | | Release date: | 2010-01-05 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein crystallography reveals a role for the FS0 cluster of Escherichia coli nitrate reductase A (NarGHI) in enzyme maturation.
J.Biol.Chem., 285, 2010
|
|
3IWW
 
 | |
2CHM
 
 | | Crystal structure of N2 substituted pyrazolo pyrimidinones - a flipped binding mode in PDE5 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[2-(BUT-3-EN-1-YLOXY)-5-(1-HYDROXYVINYL)PYRIDIN-3-YL]-3-ETHYL-2-(1-ETHYLAZETIDIN-3-YL)-1,2,6,7A-TETRAHYDRO-7H-PYRAZOLO[4,3-D]PYRIMIDIN-7-ONE, CGMP-SPECIFIC 3', ... | | Authors: | Allerton, C.M.N, Barber, C.G, Beaumont, K.C, Brown, D.G, Cole, S.M, Ellis, D, Lane, C.A.L, Maw, G.N, Mount, N.M, Rawson, D.J, Robinson, C.M, Street, S.D.A, Summerhill, N.W. | | Deposit date: | 2006-03-15 | | Release date: | 2006-06-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Novel Series of Potent and Selective Pde5 Inhibitors with Potential for High and Dose-Independent Oral Bioavailability
J.Med.Chem., 49, 2006
|
|
2Q9M
 
 | | 4-Substituted Trinems as Broad Spectrum-Lactamase Inhibitors: Structure-based Design, Synthesis and Biological Activity | | Descriptor: | (1R,4S,7AS)-1-(1-FORMYLPROP-1-EN-1-YL)-4-METHOXY-2,4,5,6,7,7A-HEXAHYDRO-1H-ISOINDOLE-3-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Plantan, I, Selic, L, Mesar, T, Stefanic Anderluh, P, Oblak, M, Prezelj, A, Hesse, L, Andrejasic, M, Vilar, M, Turk, D, Kocijan, A, Prevec, T, Vilfan, G, Kocjan, D, Copar, A, Urleb, U, Solmajer, T. | | Deposit date: | 2007-06-13 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 4-Substituted Trinems as Broad Spectrum beta-Lactamase Inhibitors: Structure-Based Design, Synthesis, and Biological Activity
J.Med.Chem., 50, 2007
|
|
2Q7Y
 
 | | Structure of the endogenous iNKT cell ligand iGb3 bound to mCD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Zajonc, D.M, Wilson, I.A, Teyton, L. | | Deposit date: | 2007-06-07 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Mouse CD1d-iGb3 Complex and its Cognate Valpha14 T Cell Receptor Suggest a Model for Dual Recognition of Foreign and Self Glycolipids.
J.Mol.Biol., 377, 2008
|
|
2R2H
 
 | | Structure of S25-2 in Complex with Ko | | Descriptor: | Fab, antibody fragment (IgG1k), heavy chain, ... | | Authors: | Brooks, C.L, Evans, S.V. | | Deposit date: | 2007-08-25 | | Release date: | 2008-12-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploration of specificity in germline monoclonal antibody recognition of a range of natural and synthetic epitopes.
J.Mol.Biol., 377, 2008
|
|
2QNV
 
 | | Crystal Structure of the Pregnane X Receptor bound to Colupulone | | Descriptor: | 3,5-dihydroxy-4,6,6-tris(3-methylbut-2-en-1-yl)-2-(2-methylpropanoyl)cyclohexa-2,4-dien-1-one, Orphan nuclear receptor PXR | | Authors: | Teotico, D.G, Bischof, J, Redinbo, M.R. | | Deposit date: | 2007-07-19 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Pregnane X Receptor bound to Colupulone
TO BE PUBLISHED
|
|
2QIQ
 
 | |
2R2E
 
 | |
2QU9
 
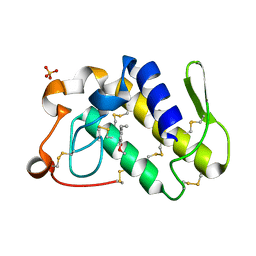 | | Crystal structure of the complex of group II phospholipase A2 with Eugenol | | Descriptor: | 2-methoxy-4-[(1E)-prop-1-en-1-yl]phenol, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Kumar, S, Vikram, G, Singh, N, Sinha, M, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2007-08-04 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of the complex of group II phospholipase A2 with Eugenol
To be Published
|
|
2QV4
 
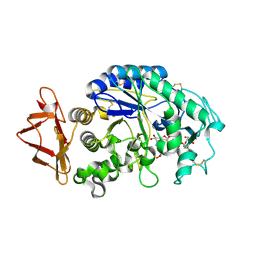 | | Human pancreatic alpha-amylase complexed with nitrite and acarbose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5R,6S)-4-{[alpha-D-glucopyranosyl-(1->4)-alpha-D-glucopyranosyl-(1->4)-alpha-D-glucopyranosyl]oxy}-5,6-dihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Williams, L.K, Maurus, R, Brayer, G.D. | | Deposit date: | 2007-08-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
2QIM
 
 | | Crystal Structure of Pathogenesis-related Protein LlPR-10.2B from yellow lupine in complex with Cytokinin | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, CALCIUM ION, GLYCEROL, ... | | Authors: | Fernandes, H.C, Pasternak, O, Bujacz, G, Bujacz, A, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2007-07-05 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Lupinus luteus pathogenesis-related protein as a reservoir for cytokinin.
J.Mol.Biol., 378, 2008
|
|
2D2C
 
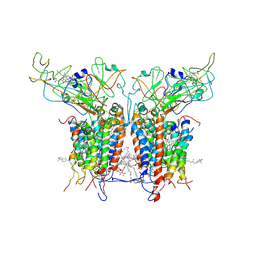 | | Crystal Structure Of Cytochrome B6F Complex with DBMIB From M. Laminosus | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 2,5-DIBROMO-3-ISOPROPYL-6-METHYLBENZO-1,4-QUINONE, Apocytochrome f, ... | | Authors: | Yan, J, Kurisu, G, Cramer, W.A. | | Deposit date: | 2005-09-07 | | Release date: | 2005-12-13 | | Last modified: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Intraprotein transfer of the quinone analogue inhibitor 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone in the cytochrome b6f complex
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2RID
 
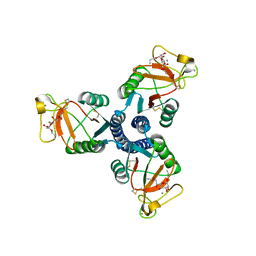 | | Crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with Allyl 7-O-carbamoyl-L-glycero-D-manno-heptopyranoside | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein D, prop-2-en-1-yl 7-O-carbamoyl-L-glycero-alpha-D-manno-heptopyranoside | | Authors: | Wang, H, Head, J, Kosma, P, Sheikh, S, McDonald, B, Smith, K, Cafarella, T, Seaton, B, Crouch, E. | | Deposit date: | 2007-10-10 | | Release date: | 2008-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recognition of heptoses and the inner core of bacterial lipopolysaccharides by surfactant protein d.
Biochemistry, 47, 2008
|
|
2E3Q
 
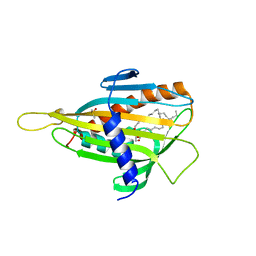 | | Crystal structure of CERT START domain in complex with C18-ceramide (P212121) | | Descriptor: | DIMETHYL SULFOXIDE, Lipid-transfer protein CERT, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)STEARAMIDE | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2E74
 
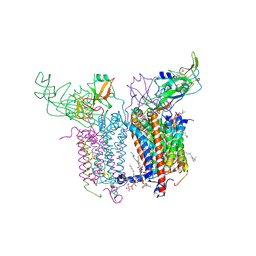 | | Crystal Structure of the Cytochrome b6f Complex from M.laminosus | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, Apocytochrome f, ... | | Authors: | Cramer, W.A, Yamashita, E, Zhang, H. | | Deposit date: | 2007-01-05 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Cytochrome b(6)f Complex: Quinone Analogue Inhibitors as Ligands of Heme c(n)
J.Mol.Biol., 370, 2007
|
|
2E3P
 
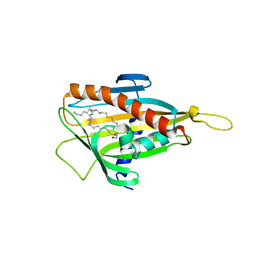 | | Crystal structure of CERT START domain in complex with C16-cearmide (P1) | | Descriptor: | Lipid-transfer protein CERT, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2E76
 
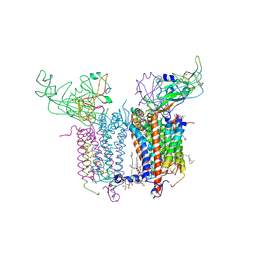 | | Crystal Structure of the Cytochrome b6f Complex with tridecyl-stigmatellin (TDS) from M.laminosus | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 8-HYDROXY-5,7-DIMETHOXY-3-METHYL-2-TRIDECYL-4H-CHROMEN-4-ONE, ... | | Authors: | Cramer, W.A, Yamashita, E, Zhang, H. | | Deposit date: | 2007-01-05 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Structure of the Cytochrome b(6)f Complex: Quinone Analogue Inhibitors as Ligands of Heme c(n)
J.Mol.Biol., 370, 2007
|
|
2E3N
 
 | | Crystal structure of CERT START domain in complex with C6-ceramide (P212121) | | Descriptor: | Lipid-transfer protein CERT, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)HEXANAMIDE | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2VAX
 
 | | Crystal structure of deacetylcephalosporin C acetyltransferase (Cephalosporin C-soak) | | Descriptor: | 4-(3-ACETOXYMETHYL-2-CARBOXY-8-OXO-5-THIA-1-AZA-BICYCLO[4.2.0]OCT-2-EN-7-YLCARBAMOYL)-1-CARBOXY-BUTYL-AMMONIUM, ACETATE ION, ACETYL-COA--DEACETYLCEPHALOSPORIN C ACETYLTRANSFERASE | | Authors: | Lejon, S, Ellis, J, Valegard, K. | | Deposit date: | 2007-09-04 | | Release date: | 2008-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Last Step in Cephalosporin C Formation Revealed: Crystal Structures of Deacetylcephalosporin C Acetyltransferase from Acremonium Chrysogenum in Complexes with Reaction Intermediates.
J.Mol.Biol., 377, 2008
|
|
2E3R
 
 | | Crystal structure of CERT START domain in complex with C18-ceramide (P1) | | Descriptor: | Lipid-transfer protein CERT, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)STEARAMIDE | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2VAV
 
 | | Crystal structure of deacetylcephalosporin C acetyltransferase (DAC-Soak) | | Descriptor: | 4-(3-ACETOXYMETHYL-2-CARBOXY-8-OXO-5-THIA-1-AZA-BICYCLO[4.2.0]OCT-2-EN-7-YLCARBAMOYL)-1-CARBOXY-BUTYL-AMMONIUM, ACETATE ION, ACETYL-COA--DEACETYLCEPHALOSPORIN C ACETYLTRANSFERASE | | Authors: | Lejon, S, Ellis, J, Valegard, K. | | Deposit date: | 2007-09-04 | | Release date: | 2008-09-23 | | Last modified: | 2018-12-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The last step in cephalosporin C formation revealed: crystal structures of deacetylcephalosporin C acetyltransferase from Acremonium chrysogenum in complexes with reaction intermediates.
J. Mol. Biol., 377, 2008
|
|
