1T14
 
 | |
2FJY
 
 | |
5DIC
 
 | |
1TWO
 
 | |
2H8V
 
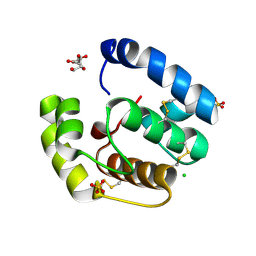 | | Structure of empty Pheromone Binding Protein ASP1 from the Honeybee Apis mellifera L | | Descriptor: | CHLORIDE ION, GLYCEROL, Pheromone-binding protein ASP1, ... | | Authors: | Pesenti, M.E, Spinelli, S, Briand, L, Pernollet, J.-C, Cambillau, C, Tegoni, M. | | Deposit date: | 2006-06-08 | | Release date: | 2007-12-11 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational Changes of the Pheromone Binding Protein ASP1 from the Honeybee Apis mellifera L upon Ligand Binding
To be Published
|
|
2GTE
 
 | | Drosophila OBP LUSH bound to attractant pheromone 11-cis-vaccenyl acetate | | Descriptor: | (Z)-OCTADEC-11-ENYL ACETATE, General odorant-binding protein lush, PHOSPHATE ION | | Authors: | Laughlin, J.D, Ha, T, Smith, D.P, Jones, D.N.M. | | Deposit date: | 2006-04-27 | | Release date: | 2007-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein
Cell(Cambridge,Mass.), 133, 2008
|
|
6OMW
 
 | |
6OPB
 
 | |
6P2E
 
 | |
7UO6
 
 | |
7VWA
 
 | |
7VW8
 
 | |
7VW9
 
 | |
6QQ4
 
 | | Odorant-binding protein dmelOBP28a from Drosophila melanogaster | | Descriptor: | General odorant-binding protein 28a, MALONIC ACID, PENTAETHYLENE GLYCOL | | Authors: | Gonzalez, D, Neiers, F, Gotthard, G, Briand, L. | | Deposit date: | 2019-02-17 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | The Drosophila odorant-binding protein 28a is involved in the detection of the floral odour ss-ionone.
Cell.Mol.Life Sci., 77, 2020
|
|
3OGN
 
 | |
3PJI
 
 | |
8BXU
 
 | | Crystal structure of Odorant Binding Protein 5 from Anopheles gambiae (AgamOBP5) with MPD (2-Methyl-2,4-pentanediol) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-ETHOXYETHANOL, Odorant binding protein, ... | | Authors: | Liggri, P.G.V, Tsitsanou, K.E, Zographos, S.E. | | Deposit date: | 2022-12-09 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structure of AgamOBP5 in complex with the natural insect repellents Carvacrol and Thymol: Crystallographic, fluorescence and thermodynamic binding studies.
Int.J.Biol.Macromol., 237, 2023
|
|
8BXW
 
 | | Crystal structure of Odorant Binding Protein 5 from Anopheles gambiae (AgamOBP5) with Carvacrol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-BUTANOL, 2-methyl-5-propan-2-yl-phenol, ... | | Authors: | Liggri, P.G.V, Tsitsanou, K.E, Zographos, S.E. | | Deposit date: | 2022-12-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of AgamOBP5 in complex with the natural insect repellents Carvacrol and Thymol: Crystallographic, fluorescence and thermodynamic binding studies.
Int.J.Biol.Macromol., 237, 2023
|
|
8BXV
 
 | | Crystal structure of Odorant Binding Protein 5 from Anopheles gambiae (AgamOBP5) with Thymol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-METHYL-2-(1-METHYLETHYL)PHENOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liggri, P.G.V, Tsitsanou, K.E, Zographos, S.E. | | Deposit date: | 2022-12-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of AgamOBP5 in complex with the natural insect repellents Carvacrol and Thymol: Crystallographic, fluorescence and thermodynamic binding studies.
Int.J.Biol.Macromol., 237, 2023
|
|
3QME
 
 | |
6MT7
 
 | |
6MTF
 
 | | D7 protein from Phlebotomus duboscqi, native | | Descriptor: | 26.7 kDa salivary protein, FRAGMENT OF TRITON X-100 | | Authors: | Andersen, J.F, Jablonka, W. | | Deposit date: | 2018-10-19 | | Release date: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Functional and structural similarities of D7 proteins in the independently-evolved salivary secretions of sand flies and mosquitoes.
Sci Rep, 9, 2019
|
|
3Q8I
 
 | |
8BY4
 
 | |
8C68
 
 | |
