6WYO
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 1 (CD1) H82F F202Y double mutant complexed with Trichostatin A | | Descriptor: | Histone deacetylase 6, POTASSIUM ION, TRICHOSTATIN A, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.30000281 Å) | | Cite: | Binding of inhibitors to active-site mutants of CD1, the enigmatic catalytic domain of histone deacetylase 6.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6WYP
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 1 (CD1) K330L mutant complexed with SAHA-BPyne | | Descriptor: | Histone deacetylase 6, N~1~-(4-{4-[(hex-5-ynoyl)amino]benzene-1-carbonyl}phenyl)-N~8~-hydroxyoctanediamide, POTASSIUM ION, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.40006351 Å) | | Cite: | Binding of inhibitors to active-site mutants of CD1, the enigmatic catalytic domain of histone deacetylase 6.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6WYQ
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 1 (CD1) K330L mutant complexed with 4-iodo-SAHA | | Descriptor: | Histone deacetylase 6, N~1~-hydroxy-N~8~-(4-iodophenyl)octanediamide, POTASSIUM ION, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.90001464 Å) | | Cite: | Binding of inhibitors to active-site mutants of CD1, the enigmatic catalytic domain of histone deacetylase 6.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6XDM
 
 | |
6XEB
 
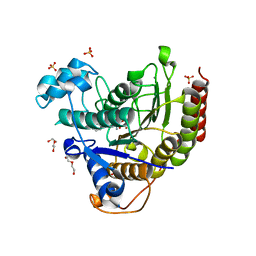 | | STRUCTURE OF HUMAN HDAC2 IN COMPLEX WITH KETONE INHIBITOR (COMPOUND E) | | Descriptor: | 5-{(1S)-7,7-dihydroxy-1-[(1-methylazetidine-3-carbonyl)amino]nonyl}-2-phenyl-1H-imidazole-4-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Clausen, D. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Development of a selective HDAC inhibitor aimed at reactivating the HIV latent reservoir.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6XEC
 
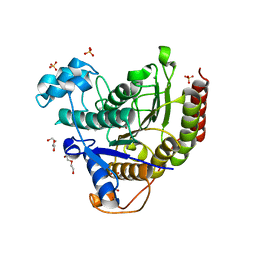 | | STRUCTURE OF HUMAN HDAC2 IN COMPLEX WITH KETONE INHIBITOR (COMPOUND O) | | Descriptor: | (1S)-N-{(1S)-1-[5-cyano-2-(4-fluorophenyl)-1H-imidazol-4-yl]-7,7-dihydroxynonyl}-6-methyl-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Clausen, D. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Development of a selective HDAC inhibitor aimed at reactivating the HIV latent reservoir.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6Z2J
 
 | | The structure of the dimeric HDAC1/MIDEAS/DNTTIP1 MiDAC deacetylase complex | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Fairall, L, Saleh, A, Ragan, T.J, Millard, C.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-05-16 | | Release date: | 2020-07-08 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The MiDAC histone deacetylase complex is essential for embryonic development and has a unique multivalent structure.
Nat Commun, 11, 2020
|
|
6Z2K
 
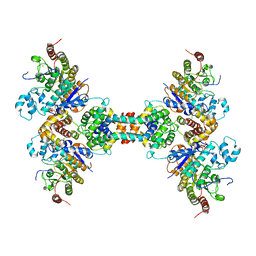 | | The structure of the tetrameric HDAC1/MIDEAS/DNTTIP1 MiDAC deacetylase complex | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Fairall, L, Saleh, A, Ragan, T.J, Millard, C.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-05-16 | | Release date: | 2020-07-08 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The MiDAC histone deacetylase complex is essential for embryonic development and has a unique multivalent structure.
Nat Commun, 11, 2020
|
|
6Z6F
 
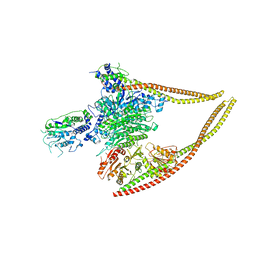 | | HDAC-PC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6H
 
 | | HDAC-DC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (8.55 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6O
 
 | | HDAC-TC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6P
 
 | | HDAC-PC-Nuc | | Descriptor: | DNA (145-MER), HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6ZW1
 
 | | X-ray structure of Danio rerio histone deacetylase 6 (HDAC6) CD2 in complex with an inhibitor SW101 | | Descriptor: | 1-[[4-(oxidanylcarbamoyl)phenyl]methyl]-3,4-dihydro-2~{H}-quinoline-6-carboxamide, GLYCEROL, Histone deacetylase 6, ... | | Authors: | Barinka, C, Motlova, L, Ustinova, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Tetrahydroquinoline-Capped Histone Deacetylase 6 Inhibitor SW-101 Ameliorates Pathological Phenotypes in a Charcot-Marie-Tooth Type 2A Mouse Model.
J.Med.Chem., 64, 2021
|
|
7AO8
 
 | | Structure of the MTA1/HDAC1/MBD2 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
7AO9
 
 | | Structure of the core MTA1/HDAC1/MBD2 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
7AOA
 
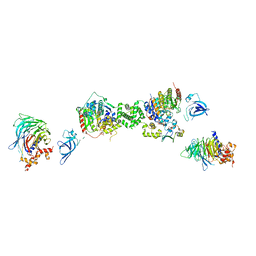 | | Structure of the extended MTA1/HDAC1/MBD2/RBBP4 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, Histone-binding protein RBBP4, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (19.4 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
7JOM
 
 | |
7JS8
 
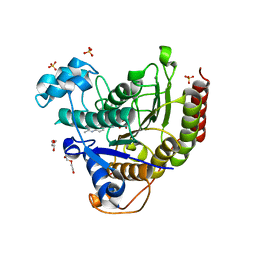 | | STRUCTURE OF HUMAN HDAC2 IN COMPLEX WITH AN ETHYL KETONE INHIBITOR CONTAINING A SPIRO-BICYCLIC GROUP (COMPOUND 22) | | Descriptor: | (1S)-N-{(1S)-7,7-dihydroxy-1-[4-(2-methylquinolin-6-yl)-1H-imidazol-2-yl]nonyl}-6-methyl-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2020-08-14 | | Release date: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (1.634 Å) | | Cite: | Discovery of Ethyl Ketone-Based Highly Selective HDACs 1, 2, 3 Inhibitors for HIV Latency Reactivation with Minimum Cellular Potency Serum Shift and Reduced hERG Activity.
J.Med.Chem., 64, 2021
|
|
7JVU
 
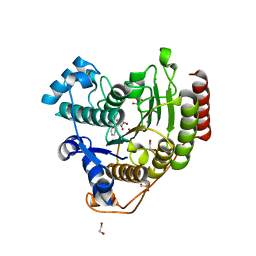 | | Crystal structure of human histone deacetylase 8 (HDAC8) I45T mutation complexed with SAHA | | Descriptor: | 1,2-ETHANEDIOL, Histone deacetylase 8, OCTANEDIOIC ACID HYDROXYAMIDE PHENYLAMIDE, ... | | Authors: | Osko, J.D, Christianson, D.W, Decroos, C, Porter, N.J, Lee, M. | | Deposit date: | 2020-08-24 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5004766 Å) | | Cite: | Structural analysis of histone deacetylase 8 mutants associated with Cornelia de Lange Syndrome spectrum disorders.
J.Struct.Biol., 213, 2020
|
|
7JVV
 
 | | Crystal structure of human histone deacetylase 8 (HDAC8) E66D/Y306F double mutation complexed with a tetrapeptide substrate | | Descriptor: | 1,2-ETHANEDIOL, ACE-ARG-HIS-ALY-ALY-MCM, GLYCEROL, ... | | Authors: | Osko, J.D, Christianson, D.W, Decroos, C, Porter, N.J, Lee, M. | | Deposit date: | 2020-08-24 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of histone deacetylase 8 mutants associated with Cornelia de Lange Syndrome spectrum disorders.
J.Struct.Biol., 213, 2020
|
|
7JVW
 
 | | Crystal structure of human histone deacetylase 8 (HDAC8) G320R mutation complexed with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Osko, J.D, Christianson, D.W, Decroos, C, Porter, N.J, Lee, M. | | Deposit date: | 2020-08-24 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.40301776 Å) | | Cite: | Structural analysis of histone deacetylase 8 mutants associated with Cornelia de Lange Syndrome spectrum disorders.
J.Struct.Biol., 213, 2020
|
|
7KBG
 
 | |
7KBH
 
 | |
7KUQ
 
 | |
7KUR
 
 | |
