8FRC
 
 | |
8FRB
 
 | | Mouse acidic mammalian chitinase, catalytic domain in complex with N,N'-diacetylchitobiose at pH 5.25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetylamino-2-deoxy-alpha-L-idopyranose, ... | | Authors: | Diaz, R.E, Fraser, J.S. | | Deposit date: | 2023-01-06 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of ligand binding and pH-specific enzymatic activity of mouse Acidic Mammalian Chitinase.
Biorxiv, 2024
|
|
8FG5
 
 | | Apo mouse acidic mammalian chitinase, catalytic domain at 100 K | | Descriptor: | Acidic mammalian chitinase, MAGNESIUM ION | | Authors: | Diaz, R.E, Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2022-12-12 | | Release date: | 2023-03-01 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural characterization of ligand binding and pH-specific enzymatic activity of mouse Acidic Mammalian Chitinase.
Biorxiv, 2024
|
|
8FR9
 
 | |
8FG7
 
 | |
8FRA
 
 | | Mouse acidic mammalian chitinase, catalytic domain in complex with diacetylchitobiose at pH 5.60 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acidic mammalian chitinase, ... | | Authors: | Diaz, R.E, Fraser, J.S. | | Deposit date: | 2023-01-06 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of ligand binding and pH-specific enzymatic activity of mouse Acidic Mammalian Chitinase.
Biorxiv, 2024
|
|
8FRG
 
 | |
8FRD
 
 | |
8GCA
 
 | | Mouse acidic mammalian chitinase, catalytic domain in complex with N,N',N''-triacetylchitotriose at pH 4.74 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acidic mammalian chitinase, ... | | Authors: | Diaz, R.E, Fraser, J.S. | | Deposit date: | 2023-03-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of ligand binding and pH-specific enzymatic activity of mouse Acidic Mammalian Chitinase.
Biorxiv, 2024
|
|
6K7Z
 
 | | Crystal structure of a GH18 chitinase from Pseudoalteromonas aurantia | | Descriptor: | GH18 chiitnase | | Authors: | Wang, Y.J, Li, P.Y, Cao, H.Y, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structural Insight Into Chitin Degradation and Thermostability of a Novel Endochitinase From the Glycoside Hydrolase Family 18.
Front Microbiol, 10, 2019
|
|
2QF8
 
 | | Crystal structure of the complex of Buffalo Secretory Glycoprotein with tetrasaccharide at 2.8A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1 | | Authors: | Singh, A.K, Jain, R, Sinha, M, Kumar, A, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-06-27 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the complex of Buffalo Secretory Glycoprotein with Tetrasaccharide at 2.8A resolution
To be Published
|
|
6KPO
 
 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris D154N/E156Q mutant in complex with fucosyl-N-acetylglucosamine-Asn | | Descriptor: | ASPARAGINE, Chitinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
6KPN
 
 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris D154N/E156Q mutant in complex with fucosyl-N-acetylglucosamine | | Descriptor: | Chitinase, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
6KST
 
 | |
6KXN
 
 | | Crystal structure of W50A mutant of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ueda, M, Shimosaka, M, Arai, R. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of CsChiL, a chitinase from Chitiniphilus shinanonensis
To be published
|
|
6KPM
 
 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in complex with L-fucose | | Descriptor: | Chitinase, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
8OVR
 
 | |
6KPL
 
 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in apo form | | Descriptor: | Chitinase, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
6KXL
 
 | | Crystal structure of the catalytic domain of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose | | Descriptor: | 1,2-ETHANEDIOL, 2-METHOXYETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ueda, M, Shimosaka, M, Arai, R. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of CsChiL, a chitinase from Chitiniphilus shinanonensis
To be published
|
|
6KXM
 
 | |
8P8S
 
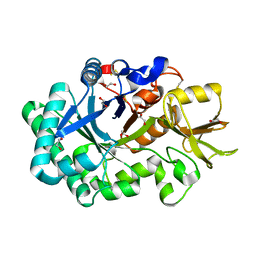 | | Recombinant Ym2 crystal structure | | Descriptor: | 1,2-ETHANEDIOL, Chitinase-like protein 4 | | Authors: | Verschueren, K.H.G, Verstraete, K, Heyndrickx, I, Smole, U, Aegerter, A, Savvides, S.N, Lambrecht, B.N. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Ym1 protein crystals promote type 2 immunity.
Elife, 12, 2024
|
|
8P8T
 
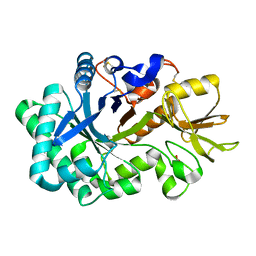 | | Ex vivo Ym2 crystal structure | | Descriptor: | Chitinase-like protein 4, GLYCEROL | | Authors: | Verschueren, K.H.G, Verstraete, K, Heyndrickx, I, Smole, U, Aegerter, H, Savvides, S.N, Lambrecht, B.N. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Ym1 protein crystals promote type 2 immunity.
Elife, 12, 2024
|
|
8P8Q
 
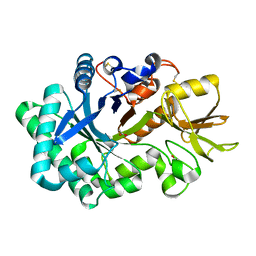 | | Recombinant Ym1 crystal structure | | Descriptor: | ACETATE ION, Chitinase-like protein 3, GLYCEROL | | Authors: | Verschueren, K.H.G, Verstraete, K, Heyndrickx, I, Aegerter, H, Smole, U, Savvides, S.N, Lambrecht, B.N. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Ym1 protein crystals promote type 2 immunity.
Elife, 12, 2024
|
|
8P8R
 
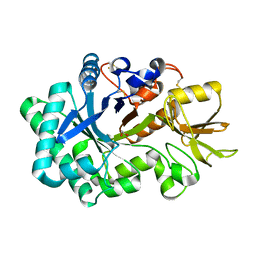 | | Ex vivo Ym1 crystal structure | | Descriptor: | 1,2-ETHANEDIOL, Chitinase-like protein 3 | | Authors: | Verschueren, K.H.G, Verstraete, K, Heyndrickx, I, Smole, U, Aegerter, H, Savvides, S.N, Lambrecht, B.N. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Ym1 protein crystals promote type 2 immunity.
Elife, 12, 2024
|
|
6LDU
 
 | |
