2LOB
 
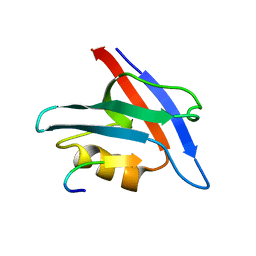 | | PDZ Domain of CAL (Cystic Fibrosis Transmembrane Regulator-Associated Ligand) | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, Golgi-associated PDZ and coiled-coil motif-containing protein | | Authors: | Piserchio, A, Fellows, A, Madden, D.R, Mierke, D.F. | | Deposit date: | 2012-01-20 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Association of the cystic fibrosis transmembrane regulator with CAL: structural features and molecular dynamics.
Biochemistry, 44, 2005
|
|
4A9G
 
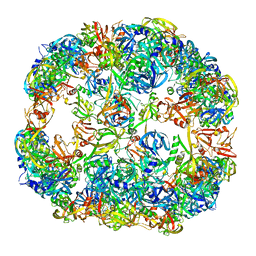 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 24-mer in complex with beta-casein | | Descriptor: | PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A8B
 
 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 12-mer in complex with lysozymes | | Descriptor: | LYSOZYME C, PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-20 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A8C
 
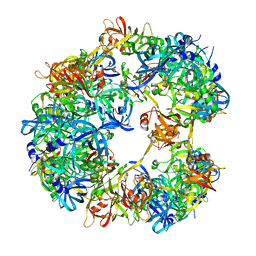 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 12-mer in complex with a binding peptide | | Descriptor: | PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-20 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A8D
 
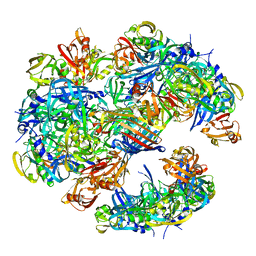 | | DegP dodecamer with bound OMP | | Descriptor: | OUTER MEMBRANE PROTEIN C, PERIPLASMIC SERINE ENDOPROTEASE DEGP | | Authors: | Malet, H, Krojer, T, Sawa, J, Schafer, E, Saibil, H.R, Ehrmann, M, Clausen, T. | | Deposit date: | 2011-11-20 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A8A
 
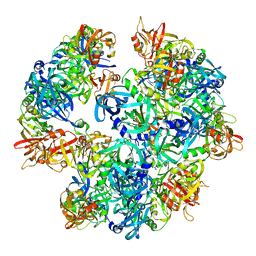 | | Asymmetric cryo-EM reconstruction of E. coli DegQ 12-mer in complex with lysozyme | | Descriptor: | LYSOZYME C, PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-20 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14.2 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
3TSZ
 
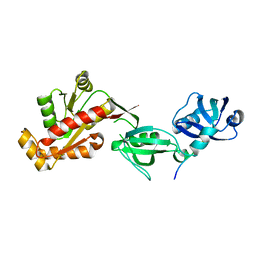 | |
3TSV
 
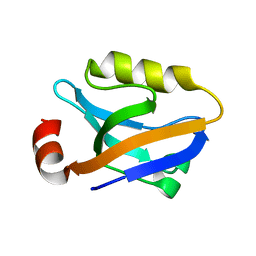 | |
3TSW
 
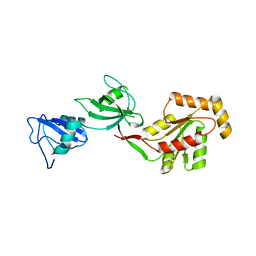 | | crystal structure of the PDZ3-SH3-GUK core module of Human ZO-1 | | Descriptor: | SULFATE ION, Tight junction protein ZO-1 | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | The Src Homology 3 Domain Is Required for Junctional Adhesion Molecule Binding to the Third PDZ Domain of the Scaffolding Protein ZO-1.
J.Biol.Chem., 286, 2011
|
|
3TMH
 
 | | Crystal structure of dual-specific A-kinase anchoring protein 2 in complex with cAMP-dependent protein kinase A type II alpha and PDZK1 | | Descriptor: | A-kinase anchor protein 10, mitochondrial, Na(+)/H(+) exchange regulatory cofactor NHE-RF3, ... | | Authors: | Sarma, G.N, Phan, R.H, Sankaran, B, Taylor, S.S. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Differential mode of D-AKAP2 to PKA isoforms: Insights from D-AKAP2 PKA RII PDZK1 ternary complex structure
To be Published
|
|
3STJ
 
 | | Crystal structure of the protease + PDZ1 domain of DegQ from Escherichia coli | | Descriptor: | Protease degQ, peptide (UNK) | | Authors: | Sawa, J, Malet, H, Krojer, T, Canellas, F, Ehrmann, M, Clausen, T. | | Deposit date: | 2011-07-11 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular adaptation of the DegQ protease to exert protein quality control in the bacterial cell envelope.
J.Biol.Chem., 286, 2011
|
|
3SUZ
 
 | | Crystal structure of Rat Mint2 PPC | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 2 | | Authors: | Shen, Y, Long, J, Yan, X, Xie, X. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Open-closed motion of Mint2 regulates APP metabolism
J Mol Cell Biol, 5, 2013
|
|
3ZRT
 
 | | Crystal structure of human PSD-95 PDZ1-2 | | Descriptor: | DISKS LARGE HOMOLOG 4 | | Authors: | Sorensen, P.L, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2011-06-19 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.398 Å) | | Cite: | A High-Affinity, Dimeric Inhibitor of Psd-95 Bivalently Interacts with Pdz1-2 and Protects Against Ischemic Brain Damage.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3SHU
 
 | | Crystal structure of ZO-1 PDZ3 | | Descriptor: | Tight junction protein ZO-1 | | Authors: | Yu, J, Pan, L, Chen, J, Yu, H, Zhang, M. | | Deposit date: | 2011-06-17 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Structure of the PDZ3-SH3-GuK Tandem of ZO-1 Suggests a Supramodular Organization of the Conserved MAGUK Family Scaffold Core
To be Published
|
|
3SHW
 
 | | Crystal structure of ZO-1 PDZ3-SH3-Guk supramodule complex with Connexin-45 peptide | | Descriptor: | Gap junction gamma-1 protein, Tight junction protein ZO-1 | | Authors: | Yu, J, Pan, L, Chen, J, Yu, H, Zhang, M. | | Deposit date: | 2011-06-17 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of the PDZ3-SH3-GuK Tandem of ZO-1 Suggests a Supramodular Organization of the Conserved MAGUK Family Scaffold Core
To be Published
|
|
3SFJ
 
 | |
2LC6
 
 | |
2LC7
 
 | |
3RL7
 
 | | Crystal structure of hDLG1-PDZ1 complexed with APC | | Descriptor: | 11-mer peptide from Adenomatous polyposis coli protein, Disks large homolog 1 | | Authors: | Zhang, Z, Li, H, Wu, G. | | Deposit date: | 2011-04-19 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the recognition of adenomatous polyposis coli by the Discs Large 1 protein.
Plos One, 6, 2011
|
|
3RL8
 
 | | Crystal structure of hDLG1-PDZ2 complexed with APC | | Descriptor: | 11-mer peptide from Adenomatous polyposis coli protein, Disks large homolog 1 | | Authors: | Zhang, Z, Li, H, Wu, G. | | Deposit date: | 2011-04-19 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for the recognition of adenomatous polyposis coli by the Discs Large 1 protein.
Plos One, 6, 2011
|
|
3AXA
 
 | | Crystal structure of afadin PDZ domain in complex with the C-terminal peptide from nectin-3 | | Descriptor: | Afadin, Nectin-3 | | Authors: | Fujiwara, Y, Goda, N, Narita, H, Satomura, K, Nakagawa, A, Sakisaka, T, Suzuki, M, Hiroaki, H. | | Deposit date: | 2011-03-31 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of afadin PDZ domain-nectin-3 complex shows the structural plasticity of the ligand-binding site.
Protein Sci., 24, 2015
|
|
3R68
 
 | | Molecular Analysis of the PDZ3 domain of PDZK1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Kocher, O, Birrane, G, Krieger, M. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Identification of the PDZ3 Domain of the Adaptor Protein PDZK1 as a Second, Physiologically Functional Binding Site for the C Terminus of the High Density Lipoprotein Receptor Scavenger Receptor Class B Type I.
J.Biol.Chem., 286, 2011
|
|
3R69
 
 | | Molecular analysis of the interaction of the HDL-receptor SR-BI with the PDZ3 domain of its adaptor protein PDZK1 | | Descriptor: | CITRIC ACID, Na(+)/H(+) exchange regulatory cofactor NHE-RF3, Scavenger receptor class B member 1 | | Authors: | Kocher, O, Birrane, G, Krieger, M. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Identification of the PDZ3 Domain of the Adaptor Protein PDZK1 as a Second, Physiologically Functional Binding Site for the C Terminus of the High Density Lipoprotein Receptor Scavenger Receptor Class B Type I.
J.Biol.Chem., 286, 2011
|
|
2LA8
 
 | |
3R0H
 
 | | Structure of INAD PDZ45 in complex with NG2 peptide | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Inactivation-no-after-potential D protein, ... | | Authors: | Wei, Z, Liu, W, Zhang, M. | | Deposit date: | 2011-03-08 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The INAD scaffold is a dynamic, redox-regulated modulator of signaling in the Drosophila eye
Cell(Cambridge,Mass.), 145, 2011
|
|
