3WKD
 
 | | Crystal structure of soluble epoxide hydrolase in complex with fragment inhibitor | | Descriptor: | Bifunctional epoxide hydrolase 2, MAGNESIUM ION, N-[2-(morpholin-4-yl)phenyl]thiophene-3-carboxamide, ... | | Authors: | Amano, Y, Yamaguchi, T, Tanabe, E. | | Deposit date: | 2013-10-18 | | Release date: | 2014-04-16 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural insights into binding of inhibitors to soluble epoxide hydrolase gained by fragment screening and X-ray crystallography.
Bioorg.Med.Chem., 22, 2014
|
|
3WZM
 
 | | ZEN lactonase mutant complex | | Descriptor: | (3S,11E)-14,16-dihydroxy-3-methyl-3,4,5,6,9,10-hexahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Zearalenone hydrolase | | Authors: | Ko, T.P, Huang, C.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure and substrate-binding mode of the mycoestrogen-detoxifying lactonase ZHD from Clonostachys rosea
RSC ADV, 4, 2014
|
|
8PI1
 
 | | Bicyclic INCYPRO Pseudomonas fluorescens esterase | | Descriptor: | Arylesterase, GLYCEROL, N-[2-[3,5-bis[2-(2-iodanylethanoylamino)ethanoyl]-1,3,5-triazinan-1-yl]-2-oxidanylidene-ethyl]-2-iodanyl-ethanamide | | Authors: | Kiehstaller, S, Pearce, N.M, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Covalent bicyclization of protein complexes yields durable quaternary structures.
Chem, 10, 2024
|
|
4WDR
 
 | | Crystal structure of haloalkane dehalogenase LinB 140A+143L+177W+211L mutant (LinB86) from Sphingobium japonicum UT26 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Degtjarik, O, Rezacova, P, Iermak, I, Chaloupkova, R, Damborsky, J, Kuta-Smatanova, I. | | Deposit date: | 2014-09-09 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of haloalkane dehalogenase LinB mutant (L177W) from Sphingobium japonicum UT26
Acs Catalysis, 2016
|
|
2PSE
 
 | |
1WOM
 
 | | Crystal structure of RsbQ | | Descriptor: | MALONIC ACID, S-1,2-PROPANEDIOL, Sigma factor sigB regulation protein rsbQ | | Authors: | Kaneko, T, Kumasaka, T, Tanaka, N. | | Deposit date: | 2004-08-21 | | Release date: | 2005-02-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis
Protein Sci., 14, 2005
|
|
4Y2X
 
 | | Structure of soluble epoxide hydrolase in complex with 2-({[2-(adamantan-1-yl)ethyl]amino}methyl)phenol | | Descriptor: | 2-[({2-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]dec-1-yl]ethyl}amino)methyl]phenol, Bifunctional epoxide hydrolase 2, MAGNESIUM ION | | Authors: | Amano, Y, Yamaguchi, T. | | Deposit date: | 2015-02-10 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of N-ethylmethylamine as a novel scaffold for inhibitors of soluble epoxide hydrolase by crystallographic fragment screening
Bioorg.Med.Chem., 23, 2015
|
|
6YNX
 
 | | Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - Fo-subcomplex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kock Flygaard, R, Muhleip, A, Amunts, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Type III ATP synthase is a symmetry-deviated dimer that induces membrane curvature through tetramerization.
Nat Commun, 11, 2020
|
|
3RKS
 
 | | Crystal Structure of the Manihot esculenta Hydroxynitrile Lyase (MeHNL) K176P mutant | | Descriptor: | GLYCEROL, Hydroxynitrilase | | Authors: | Cielo, C.B.C, Yamane, T, Asano, Y, Dadashipour, M, Suzuki, A, Mizushima, T, Komeda, H. | | Deposit date: | 2011-04-18 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic Studies of Manihot esculenta hydroxynitrile lyase Lysine-to-Proline mutants
To be Published
|
|
2VAV
 
 | | Crystal structure of deacetylcephalosporin C acetyltransferase (DAC-Soak) | | Descriptor: | 4-(3-ACETOXYMETHYL-2-CARBOXY-8-OXO-5-THIA-1-AZA-BICYCLO[4.2.0]OCT-2-EN-7-YLCARBAMOYL)-1-CARBOXY-BUTYL-AMMONIUM, ACETATE ION, ACETYL-COA--DEACETYLCEPHALOSPORIN C ACETYLTRANSFERASE | | Authors: | Lejon, S, Ellis, J, Valegard, K. | | Deposit date: | 2007-09-04 | | Release date: | 2008-09-23 | | Last modified: | 2018-12-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The last step in cephalosporin C formation revealed: crystal structures of deacetylcephalosporin C acetyltransferase from Acremonium chrysogenum in complexes with reaction intermediates.
J. Mol. Biol., 377, 2008
|
|
5IE7
 
 | | Crystal structure of a lactonase double mutant in complex with substrate b | | Descriptor: | (3S,7S,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, Zearalenone hydrolase | | Authors: | Zheng, Y.Y, Xu, Z.X, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enhanced alph-Zearalenol Hydrolyzing Activity of a Mycoestrogen-Detoxifying Lactonase by Structure-Based Engineering
Acs Catalysis, 6, 2016
|
|
5XM6
 
 | | the overall structure of VrEH2 | | Descriptor: | Epoxide hydrolase | | Authors: | Li, F.L, Kong, X.D, Yu, H.L, Shang, Y.P, Zhou, J.H, Xu, J.H. | | Deposit date: | 2017-05-12 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Regioselectivity Engineering of Epoxide Hydrolase: Near-Perfect Enantioconvergence through a Single Site Mutation
Acs Catalysis, 8, 2018
|
|
3DQZ
 
 | | Structure of the hydroxynitrile lyase from Arabidopsis thaliana | | Descriptor: | Alpha-hydroxynitrile lyase-like protein, CHLORIDE ION | | Authors: | Andexer, J, Staunig, N, Gruber, K. | | Deposit date: | 2008-07-10 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Hydroxynitrile lyases with alpha / beta-hydrolase fold: two enzymes with almost identical 3D structures but opposite enantioselectivities and different reaction mechanisms
Chembiochem, 13, 2012
|
|
5AKG
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 5-(2-thienyl)-1,3,4-thiadiazol-2-amine, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-03 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
1EX9
 
 | | CRYSTAL STRUCTURE OF THE PSEUDOMONAS AERUGINOSA LIPASE COMPLEXED WITH RC-(RP,SP)-1,2-DIOCTYLCARBAMOYL-GLYCERO-3-O-OCTYLPHOSPHONATE | | Descriptor: | CALCIUM ION, LACTONIZING LIPASE, OCTYL-PHOSPHINIC ACID 1,2-BIS-OCTYLCARBAMOYLOXY-ETHYL ESTER | | Authors: | Nardini, M, Lang, D.A, Liebeton, K, Jaeger, K.-E, Dijkstra, B.W. | | Deposit date: | 2000-05-02 | | Release date: | 2000-10-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of pseudomonas aeruginosa lipase in the open conformation. The prototype for family I.1 of bacterial lipases.
J.Biol.Chem., 275, 2000
|
|
6XTC
 
 | |
3WWO
 
 | | S-selective hydroxynitrile lyase from Baliospermum montanum (apo1) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (S)-hydroxynitrile lyase, CALCIUM ION | | Authors: | Nakano, S, Dadashipour, M, Asano, Y. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and functional analysis of hydroxynitrile lyase from Baliospermum montanum with crystal structure, molecular dynamics and enzyme kinetics
Biochim.Biophys.Acta, 1844, 2014
|
|
4HAI
 
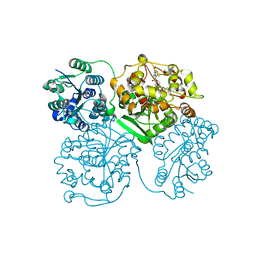 | | Crystal structure of human soluble epoxide hydrolase complexed with N-cycloheptyl-1-(mesitylsulfonyl)piperidine-4-carboxamide. | | Descriptor: | Bifunctional epoxide hydrolase 2, MAGNESIUM ION, N-cycloheptyl-1-[(2,4,6-trimethylphenyl)sulfonyl]piperidine-4-carboxamide, ... | | Authors: | Pecic, S, Pakhomova, S, Newcomer, M.E, Morisseau, C, Hammock, B.D, Zhu, Z, Deng, S. | | Deposit date: | 2012-09-26 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Synthesis and structure-activity relationship of piperidine-derived non-urea soluble epoxide hydrolase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5ALS
 
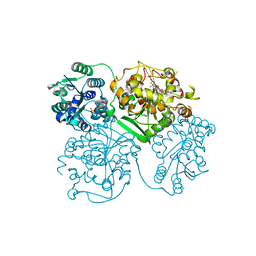 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | BIFUNCTIONAL EPOXIDE HYDROLASE 2, N-CYCLOPENTYL-2-[4-(TRIFLUOROMETHYL)PHENYL]-3H-IMIDAZO[4,5-B]PYRIDINE-7-SULFONAMIDE, SULFATE ION | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-08 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
1WPR
 
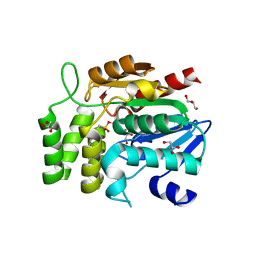 | | Crystal structure of RsbQ inhibited by PMSF | | Descriptor: | GLYCEROL, Sigma factor sigB regulation protein rsbQ, phenylmethanesulfonic acid | | Authors: | Kaneko, T, Tanaka, N, Kumasaka, T. | | Deposit date: | 2004-09-11 | | Release date: | 2005-02-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis
Protein Sci., 14, 2005
|
|
3PDC
 
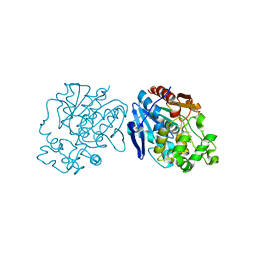 | |
1BEE
 
 | | HALOALKANE DEHALOGENASE MUTANT WITH TRP 175 REPLACED BY TYR | | Descriptor: | HALOALKANE DEHALOGENASE | | Authors: | Ridder, I.S, Vos, G.J, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1998-05-13 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Kinetic analysis and X-ray structure of haloalkane dehalogenase with a modified halide-binding site.
Biochemistry, 37, 1998
|
|
8B6Q
 
 | | X-ray structure of the haloalkane dehalogenase HaloTag7 with an insertion of Calmodulin-M13 fusion at position 154-156 that mimic the structure of CaProLa, an calcium gated protein labeling technology | | Descriptor: | CALCIUM ION, CHLORIDE ION, Haloalkane dehalogenase,Calmodulin-1,Haloalkane dehalogenase,Calmodulin-1,M13 peptide | | Authors: | Tarnawski, M, Johnsson, K, Hiblot, J. | | Deposit date: | 2022-09-27 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure of the haloalkane dehalogenase HaloTag7 with an insertion of Calmodulin-M13 fusion at position 154-156 that mimic the structure of CaProLa, an calcium gated protein labeling technology
To Be Published
|
|
6I5E
 
 | | X-ray structure of apo human soluble Epoxide Hydrolase C-terminal Domain (hsEH CTD) | | Descriptor: | Bifunctional epoxide hydrolase 2 | | Authors: | Abis, G, Kopec, J, Yue, W.W, Conte, M.R. | | Deposit date: | 2018-11-13 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 15-deoxy-Delta12,14-Prostaglandin J2inhibits human soluble epoxide hydrolase by a dual orthosteric and allosteric mechanism.
Commun Biol, 2, 2019
|
|
4YK7
 
 | |
