1U2E
 
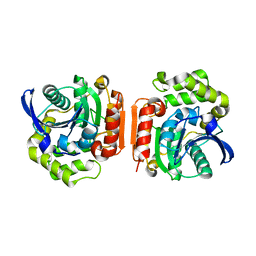 | | Crystal Structure of the C-C bond hydrolase MhpC | | Descriptor: | 2-hydroxy-6-ketonona-2,4-dienedioic acid hydrolase, CHLORIDE ION | | Authors: | Montgomery, M.G, Dunn, G, Mohammed, F, Robertson, T, Garcia, J.-L, Coker, A, Bugg, T.D.H, Wood, S.P. | | Deposit date: | 2004-07-19 | | Release date: | 2005-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the C-C Bond Hydrolase MhpC Provides Insights into its Catalytic Mechanism
J.Mol.Biol., 346, 2005
|
|
1WPR
 
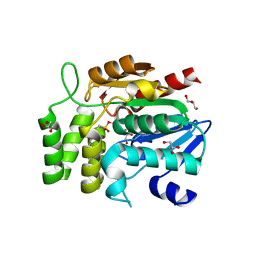 | | Crystal structure of RsbQ inhibited by PMSF | | Descriptor: | GLYCEROL, Sigma factor sigB regulation protein rsbQ, phenylmethanesulfonic acid | | Authors: | Kaneko, T, Tanaka, N, Kumasaka, T. | | Deposit date: | 2004-09-11 | | Release date: | 2005-02-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis
Protein Sci., 14, 2005
|
|
1WOM
 
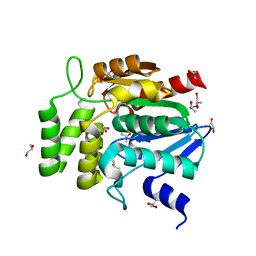 | | Crystal structure of RsbQ | | Descriptor: | MALONIC ACID, S-1,2-PROPANEDIOL, Sigma factor sigB regulation protein rsbQ | | Authors: | Kaneko, T, Kumasaka, T, Tanaka, N. | | Deposit date: | 2004-08-21 | | Release date: | 2005-02-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis
Protein Sci., 14, 2005
|
|
1UK6
 
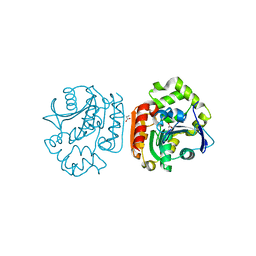 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with propionate | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, PROPANOIC ACID | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1UK8
 
 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with n-valerate | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, PENTANOIC ACID | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1UK7
 
 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with n-butyrate | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, butanoic acid | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1UKA
 
 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with (S)-2-methylbutyrate | | Descriptor: | 2-METHYLBUTANOIC ACID, 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1UKB
 
 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with benzoate | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, BENZOIC ACID | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1UK9
 
 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with isovalerate | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, ISOVALERIC ACID | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1WM1
 
 | | Crystal Structure of Prolyl Aminopeptidase, Complex with Pro-TBODA | | Descriptor: | (5-TERT-BUTYL-1,3,4-OXADIAZOL-2-YL)[(2R)-PYRROLIDIN-2-YL]METHANONE, Proline iminopeptidase | | Authors: | Nakajima, Y, Inoue, T, Ito, K, Tozaka, T, Hatakeyama, S, Tanaka, N, Nakamura, K.T, Yoshimoto, T. | | Deposit date: | 2004-07-01 | | Release date: | 2004-07-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel inhibitor for prolyl aminopeptidase from Serratia marcescens and studies on the mechanism of substrate recognition of the enzyme using the inhibitor
ARCH.BIOCHEM.BIOPHYS., 416, 2003
|
|
1VA4
 
 | | Pseudomonas fluorescens aryl esterase | | Descriptor: | Arylesterase, GLYCEROL | | Authors: | Cheeseman, J.D, Tocilj, A, Park, S, Schrag, J.D, Kazlauskas, R.J. | | Deposit date: | 2004-02-11 | | Release date: | 2004-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Structure of an aryl esterase from Pseudomonas fluorescens.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1SCQ
 
 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis in complex with acetonecyanohydrin | | Descriptor: | (S)-acetone-cyanohydrin lyase, 2-HYDROXY-2-METHYLPROPANENITRILE, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1SC9
 
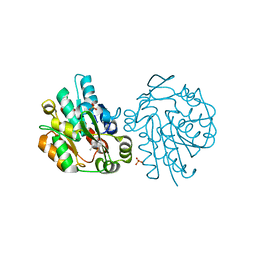 | | Hydroxynitrile Lyase from Hevea brasiliensis in complex with the natural substrate acetone cyanohydrin | | Descriptor: | (S)-acetone-cyanohydrin lyase, 2-HYDROXY-2-METHYLPROPANENITRILE, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1SCI
 
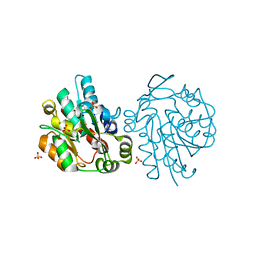 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis | | Descriptor: | (S)-acetone-cyanohydrin lyase, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1SCK
 
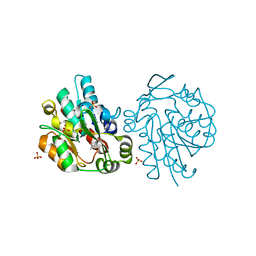 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis in complex with acetone | | Descriptor: | (S)-acetone-cyanohydrin lyase, ACETONE, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1S8O
 
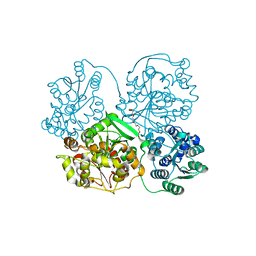 | | Human soluble Epoxide Hydrolase | | Descriptor: | HEXAETHYLENE GLYCOL, epoxide hydrolase 2, cytoplasmic | | Authors: | Gomez, G.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 2004-02-03 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human epoxide hydrolase reveals mechanistic inferences on bifunctional catalysis in epoxide and phosphate ester hydrolysis
Biochemistry, 43, 2004
|
|
1VJ5
 
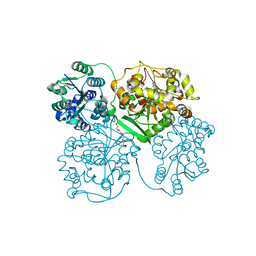 | | Human soluble Epoxide Hydrolase- N-cyclohexyl-N'-(4-iodophenyl)urea complex | | Descriptor: | HEXAETHYLENE GLYCOL, MAGNESIUM ION, N-CYCLOHEXYL-N'-(4-IODOPHENYL)UREA, ... | | Authors: | Gomez, G.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 2004-02-03 | | Release date: | 2004-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of human epoxide hydrolase reveals mechanistic inferences on bifunctional catalysis in epoxide and phosphate ester hydrolysis
Biochemistry, 43, 2004
|
|
1HKH
 
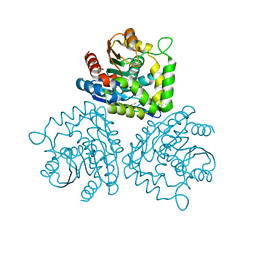 | |
1HL7
 
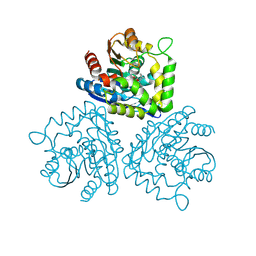 | | Gamma lactamase from an Aureobacterium species in complex with 3a,4,7,7a-tetrahydro-benzo [1,3] dioxol-2-one | | Descriptor: | 3A,4,7,7A-TETRAHYDRO-BENZO [1,3] DIOXOL-2-ONE, GAMMA LACTAMASE | | Authors: | Line, K, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2003-03-14 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Crystal Structure of a (-)Gamma-Lactamase from an Aureobacterium Species Reveals a Tetrahedral Intermediate in the Active Site
J.Mol.Biol., 338, 2004
|
|
1Q0Z
 
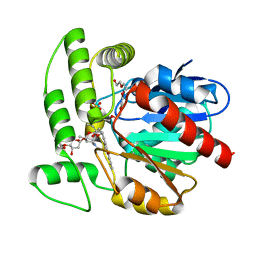 | | Crystal structure of aclacinomycin methylesterase (RdmC) with bound product analogue, 10-decarboxymethylaclacinomycin A (DcmA) | | Descriptor: | 10-DECARBOXYMETHYLACLACINOMYCIN A (DCMAA), PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Jansson, A, Niemi, J, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-07-18 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of aclacinomycin methylesterase with bound product analogues: implications for anthracycline recognition and mechanism.
J.Biol.Chem., 278, 2003
|
|
1Q0R
 
 | | Crystal structure of aclacinomycin methylesterase (RdmC) with bound product analogue, 10-decarboxymethylaclacinomycin T (DcmaT) | | Descriptor: | 10-DECARBOXYMETHYLACLACINOMYCIN T (DCMAT), PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Jansson, A, Niemi, J, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-07-17 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of aclacinomycin methylesterase with bound product analogues: implications for anthracycline recognition and mechanism.
J.Biol.Chem., 278, 2003
|
|
1MJ5
 
 | | LINB (haloalkane dehalogenase) from sphingomonas paucimobilis UT26 at atomic resolution | | Descriptor: | 1,3,4,6-tetrachloro-1,4-cyclohexadiene hydrolase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Oakley, A.J, Damborsky, J, Wilce, M.C. | | Deposit date: | 2002-08-27 | | Release date: | 2003-08-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal structure of haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26 at 0.95 A resolution: dynamics of catalytic residues.
Biochemistry, 43, 2004
|
|
1K63
 
 | | Complex of hydrolytic haloalkane dehalogenase linb from sphingomonas paucimobilis with UT26 2-BROMO-2-PROPENE-1-OL at 1.8A resolution | | Descriptor: | 1,3,4,6-tetrachloro-1,4-cyclohexadiene hydrolase, 2-BROMO-2-PROPENE-1-OL, BROMIDE ION, ... | | Authors: | Streltsov, V.A, Damborsky, J, Wilce, M.C.J. | | Deposit date: | 2001-10-15 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates
Biochemistry, 42, 2003
|
|
1K5P
 
 | | Hydrolytic haloalkane dehalogenase LINB from sphingomonas paucimobilis UT26 at 1.8A resolution | | Descriptor: | 1,3,4,6-tetrachloro-1,4-cyclohexadiene hydrolase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Streltsov, V.A, Damborsky, J, Wilce, M.C.J. | | Deposit date: | 2001-10-12 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates
Biochemistry, 42, 2003
|
|
1K6E
 
 | | COMPLEX OF HYDROLYTIC HALOALKANE DEHALOGENASE LINB FROM SPHINGOMONAS PAUCIMOBILIS UT26 WITH 1,2-PROPANEDIOL (PRODUCT OF DEHALOGENATION OF 1,2-DIBROMOPROPANE) AT 1.85A | | Descriptor: | 1-BROMOPROPANE-2-OL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Streltsov, V.A, Prokop, Z, Damborsky, J, Nagata, Y, Oakley, A, Wilce, M.C.J. | | Deposit date: | 2001-10-16 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates.
Biochemistry, 42, 2003
|
|
