3V1N
 
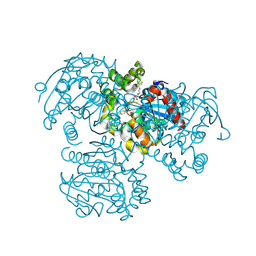 | | Crystal Structure of the H265Q mutant of a C-C hydrolase, BphD from Burkholderia xenovorans LB400, after exposure to its substrate HOPDA | | Descriptor: | (3E)-2,6-DIOXO-6-PHENYLHEX-3-ENOATE, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, BENZOIC ACID, ... | | Authors: | Ghosh, S, Bolin, J.T. | | Deposit date: | 2011-12-09 | | Release date: | 2012-03-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Identification of an Acyl-Enzyme Intermediate in a meta-Cleavage Product Hydrolase Reveals the Versatility of the Catalytic Triad.
J.Am.Chem.Soc., 134, 2012
|
|
3V1K
 
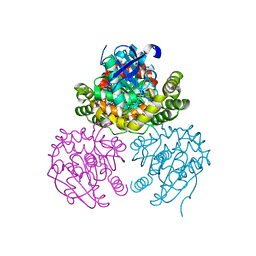 | |
3VVL
 
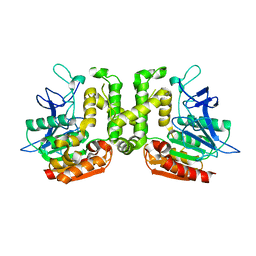 | | Crystal structure of L-serine-O-acetyltransferase found in D-cycloserine biosynthetic pathway | | Descriptor: | Homoserine O-acetyltransferase | | Authors: | Oda, K, Matoba, Y, Kumagai, T, Noda, M, Sugiyama, M. | | Deposit date: | 2012-07-26 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystallographic study to determine the substrate specificity of an L-serine-acetylating enzyme found in the D-cycloserine biosynthetic pathway
J.Bacteriol., 195, 2013
|
|
3VVM
 
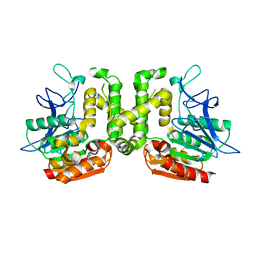 | | Crystal structure of G52A-P55G mutant of L-serine-O-acetyltransferase found in D-cycloserine biosynthetic pathway | | Descriptor: | Homoserine O-acetyltransferase | | Authors: | Oda, K, Matoba, Y, Kumagai, T, Noda, M, Sugiyama, M. | | Deposit date: | 2012-07-26 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic study to determine the substrate specificity of an L-serine-acetylating enzyme found in the D-cycloserine biosynthetic pathway
J.Bacteriol., 195, 2013
|
|
3WI7
 
 | | Crystal Structure of the Novel Haloalkane Dehalogenase DatA from Agrobacterium tumefaciens C58 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Haloalkane dehalogenase | | Authors: | Guan, L.J, Yabuki, H, Okai, M, Ohtsuka, J, Tanokura, M. | | Deposit date: | 2013-09-06 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the novel haloalkane dehalogenase DatA from Agrobacterium tumefaciens C58 reveals a special halide-stabilizing pair and enantioselectivity mechanism.
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3WIB
 
 | | Crystal structure of Y109W Mutant Haloalkane Dehalogenase DatA from Agrobacterium tumefaciens C58 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Haloalkane dehalogenase | | Authors: | Guan, L.J, Yabuki, H, Okai, M, Ohtsuka, J, Tanokura, M. | | Deposit date: | 2013-09-09 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the novel haloalkane dehalogenase DatA from Agrobacterium tumefaciens C58 reveals a special halide-stabilizing pair and enantioselectivity mechanism.
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3W9U
 
 | | Crystal structure of Lipk107 | | Descriptor: | Putative lipase | | Authors: | Yuan, Y.A. | | Deposit date: | 2013-04-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Lipk107
To be Published
|
|
3WWO
 
 | | S-selective hydroxynitrile lyase from Baliospermum montanum (apo1) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (S)-hydroxynitrile lyase, CALCIUM ION | | Authors: | Nakano, S, Dadashipour, M, Asano, Y. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and functional analysis of hydroxynitrile lyase from Baliospermum montanum with crystal structure, molecular dynamics and enzyme kinetics
Biochim.Biophys.Acta, 1844, 2014
|
|
3WMR
 
 | | Crystal structure of VinJ | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, Proline iminopeptidase | | Authors: | Shinohara, Y, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2013-11-22 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of the amidohydrolase VinJ shows a unique hydrophobic tunnel for its interaction with polyketide substrates
Febs Lett., 588, 2014
|
|
3WWP
 
 | | S-selective hydroxynitrile lyase from Baliospermum montanum (apo2) | | Descriptor: | (S)-hydroxynitrile lyase, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Nakano, S, Dadashipour, M, Asano, Y. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of hydroxynitrile lyase from Baliospermum montanum with crystal structure, molecular dynamics and enzyme kinetics
Biochim.Biophys.Acta, 1844, 2014
|
|
3WZL
 
 | | ZEN lactonase | | Descriptor: | Zearalenone hydrolase | | Authors: | Ko, T.P, Huang, C.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and substrate-binding mode of the mycoestrogen-detoxifying lactonase ZHD from Clonostachys rosea
RSC ADV, 4, 2014
|
|
3WZM
 
 | | ZEN lactonase mutant complex | | Descriptor: | (3S,11E)-14,16-dihydroxy-3-methyl-3,4,5,6,9,10-hexahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Zearalenone hydrolase | | Authors: | Ko, T.P, Huang, C.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure and substrate-binding mode of the mycoestrogen-detoxifying lactonase ZHD from Clonostachys rosea
RSC ADV, 4, 2014
|
|
3YAS
 
 | | HYDROXYNITRILE LYASE COMPLEXED WITH ACETONE | | Descriptor: | ACETONE, PROTEIN (HYDROXYNITRILE LYASE), SULFATE ION | | Authors: | Zuegg, J, Wagner, U.G, Gugganig, M, Kratky, C. | | Deposit date: | 1999-03-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
Protein Sci., 8, 1999
|
|
3X3H
 
 | | Crystal Structure of the Manihot esculenta Hydroxynitrile Lyase (MeHNL) 3KP (K176P, K199P, K224P) triple mutant | | Descriptor: | (S)-hydroxynitrile lyase | | Authors: | Cielo, C.B.C, Yamane, T, Asano, Y, Dadashipour, M, Suzuki, A, Mizushima, T, Komeda, H, Okazaki, S. | | Deposit date: | 2015-01-21 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystallographic Studies of Manihot esculenta hydroxynitrile lyase Lysine-to-Proline mutants
TO BE PUBLISHED
|
|
1U2E
 
 | | Crystal Structure of the C-C bond hydrolase MhpC | | Descriptor: | 2-hydroxy-6-ketonona-2,4-dienedioic acid hydrolase, CHLORIDE ION | | Authors: | Montgomery, M.G, Dunn, G, Mohammed, F, Robertson, T, Garcia, J.-L, Coker, A, Bugg, T.D.H, Wood, S.P. | | Deposit date: | 2004-07-19 | | Release date: | 2005-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the C-C Bond Hydrolase MhpC Provides Insights into its Catalytic Mechanism
J.Mol.Biol., 346, 2005
|
|
1UK6
 
 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with propionate | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, PROPANOIC ACID | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1UK8
 
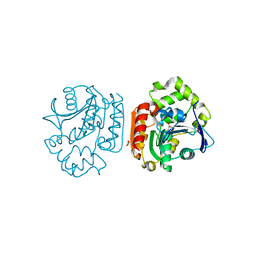 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with n-valerate | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, PENTANOIC ACID | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1UK7
 
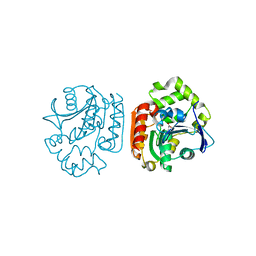 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with n-butyrate | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, butanoic acid | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1UKA
 
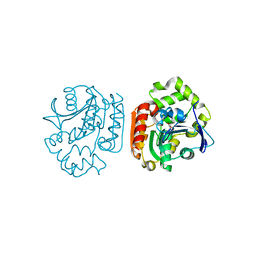 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with (S)-2-methylbutyrate | | Descriptor: | 2-METHYLBUTANOIC ACID, 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1UKB
 
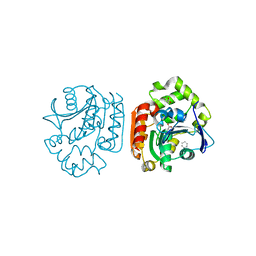 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with benzoate | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, BENZOIC ACID | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1UK9
 
 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with isovalerate | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, ISOVALERIC ACID | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1VA4
 
 | | Pseudomonas fluorescens aryl esterase | | Descriptor: | Arylesterase, GLYCEROL | | Authors: | Cheeseman, J.D, Tocilj, A, Park, S, Schrag, J.D, Kazlauskas, R.J. | | Deposit date: | 2004-02-11 | | Release date: | 2004-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Structure of an aryl esterase from Pseudomonas fluorescens.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1WM1
 
 | | Crystal Structure of Prolyl Aminopeptidase, Complex with Pro-TBODA | | Descriptor: | (5-TERT-BUTYL-1,3,4-OXADIAZOL-2-YL)[(2R)-PYRROLIDIN-2-YL]METHANONE, Proline iminopeptidase | | Authors: | Nakajima, Y, Inoue, T, Ito, K, Tozaka, T, Hatakeyama, S, Tanaka, N, Nakamura, K.T, Yoshimoto, T. | | Deposit date: | 2004-07-01 | | Release date: | 2004-07-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel inhibitor for prolyl aminopeptidase from Serratia marcescens and studies on the mechanism of substrate recognition of the enzyme using the inhibitor
ARCH.BIOCHEM.BIOPHYS., 416, 2003
|
|
1WPR
 
 | | Crystal structure of RsbQ inhibited by PMSF | | Descriptor: | GLYCEROL, Sigma factor sigB regulation protein rsbQ, phenylmethanesulfonic acid | | Authors: | Kaneko, T, Tanaka, N, Kumasaka, T. | | Deposit date: | 2004-09-11 | | Release date: | 2005-02-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis
Protein Sci., 14, 2005
|
|
1X2E
 
 | | The crystal structure of prolyl aminopeptidase complexed with Ala-TBODA | | Descriptor: | (2S)-2-AMINO-1-(5-TERT-BUTYL-1,3,4-OXADIAZOL-2-YL)PROPAN-1-ONE, Proline iminopeptidase | | Authors: | Nakajima, Y, Ito, K, Sakata, M, Xu, Y, Matsubara, F, Hatakeyama, S, Yoshimoto, T. | | Deposit date: | 2005-04-22 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Unusual extra space at the active site and high activity for acetylated hydroxyproline of prolyl aminopeptidase from Serratia marcescens
J.Bacteriol., 188, 2006
|
|
