2HHT
 
 | | C:O6-methyl-guanine pair in the polymerase-2 basepair position | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*AP*GP*CP*TP*CP*G)-3', 5'-D(*GP*TP*AP*CP*(6OG)P*AP*GP*CP*TP*GP*AP*TP*CP*GP*CP*A)-3', DNA polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HHW
 
 | | ddTTP:O6-methyl-guanine pair in the polymerase active site, in the closed conformation | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*AP*TP*(6OG)P*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*(DDG))-3', ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HHS
 
 | | O6-methyl:C pair in the polymerase-10 basepair position | | Descriptor: | 5'-D(*CP*AP*TP*(6OG)P*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*GP*CP*AP*TP*GP*A)-3', DNA polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HHQ
 
 | | O6-methyl-guanine:T pair in the polymerase-10 basepair position | | Descriptor: | 5'-D(*CP*AP*TP*(6OG)P*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*GP*TP*AP*TP*GP*A)-3', DNA polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HHU
 
 | | C:O6-methyl-guanine in the polymerase postinsertion site (-1 basepair position) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*GP*CP*GP*AP*TP*CP*AP*GP*CP*CP*C)-3', 5'-D(*GP*TP*AP*CP*(6OG)P*GP*GP*CP*TP*GP*AP*TP*CP*GP*CP*A)-3', ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HHV
 
 | | T:O6-methyl-guanine in the polymerase-2 basepair position | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*AP*GP*CP*TP*TP*G)-3', 5'-D(*GP*TP*AP*CP*(6OG)P*AP*GP*CP*TP*GP*AP*TP*CP*GP*CP*A)-3', DNA Polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HHX
 
 | | O6-methyl-guanine in the polymerase template preinsertion site | | Descriptor: | 5'-D(*CP*AP*TP*(6OG)P*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*G)-3', DNA Polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HVH
 
 | | ddCTP:O6MeG pair in the polymerase active site (0 position) | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*CP*A*TP*(6OG)P*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*(DDG))-3', ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-07-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HVI
 
 | | ddCTP:G pair in the polymerase active site (0 position) | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*C*AP*TP*GP*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*(DDG))-3', ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-07-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HW3
 
 | | T:O6-methyl-guanine pair in the polymerase postinsertion site (-1 basepair position) | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*AP*GP*CP*TP*T)-3', 5'-D(*GP*TP*A*CP*(6OG)P*AP*GP*CP*TP*GP*AP*TP*CP*GP*CP*A)-3', DNA Polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-07-31 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3EYZ
 
 | | Cocrystal structure of Bacillus fragment DNA polymerase I with duplex DNA (open form) | | Descriptor: | 5'-D(*DAP*DTP*DGP*DCP*DGP*DAP*DGP*DTP*DCP*DAP*DGP*DGP*DA)-3', 5'-D(*DCP*DCP*DTP*DGP*DAP*DCP*DTP*DCP*DGP*DC)-3', DNA polymerase I, ... | | Authors: | Warren, J.J, Wu, E.Y, Golosov, A.A, Karplus, M, Beese, L.S. | | Deposit date: | 2008-10-22 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Mechanism of the Translocation Step in DNA Replication by DNA Polymerase I: A Computer Simulation Analysis.
Structure, 18, 2010
|
|
3EZ5
 
 | | Cocrystal structure of Bacillus fragment DNA polymerase I with duplex DNA , dCTP, and zinc (closed form). | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-TRIPHOSPHATE, 5'-D(*DAP*DTP*DTP*DCP*DGP*DAP*DGP*DTP*DCP*DAP*DGP*DG)-3', 5'-D(*DCP*DCP*DTP*DGP*DAP*DCP*DTP*DCP*DG)-3', ... | | Authors: | Warren, J.J, Wu, E.Y, Golosov, A.A, Karplus, M, Beese, L.S. | | Deposit date: | 2008-10-22 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Mechanism of the Translocation Step in DNA Replication by DNA Polymerase I: A Computer Simulation Analysis.
Structure, 18, 2010
|
|
3HP6
 
 | |
3HPO
 
 | |
3HT3
 
 | |
3LWL
 
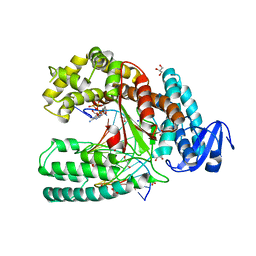 | | Structure of Klenow fragment of Taq polymerase in complex with an abasic site | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, ACETATE ION, DNA (5'-D(*AP*AP*AP*(3DR)P*TP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Replication through an abasic DNA lesion: structural basis for adenine selectivity
Embo J., 29, 2010
|
|
3LWM
 
 | | Structure of the large fragment of thermus aquaticus DNA polymerase I in complex with a blunt-ended DNA and ddATP | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, ACETATE ION, DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(2DA))-3'), ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.186 Å) | | Cite: | Replication through an abasic DNA lesion: structural basis for adenine selectivity
Embo J., 29, 2010
|
|
3M8R
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus aquaticus in a closed ternary complex with trapped 4'-ethylated dTTP | | Descriptor: | 4'-ethylthymidine 5'-(tetrahydrogen triphosphate), ACETATE ION, DNA (5'-D(*AP*AP*AP*AP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Diederichs, K, Marx, A, Betz, K. | | Deposit date: | 2010-03-19 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of DNA polymerases caught processing size-augmented nucleotide probes.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3M8S
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus aquaticus in a closed ternary complex with trapped 4'-methylated dTTP | | Descriptor: | 4'-methylthymidine 5'-(tetrahydrogen triphosphate), ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Diederichs, K, Marx, A, Betz, K. | | Deposit date: | 2010-03-19 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of DNA polymerases caught processing size-augmented nucleotide probes.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2XO7
 
 | | Crystal structure of a dA:O-allylhydroxylamine-dC basepair in complex with fragment DNA polymerase I from Bacillus stearothermophilus | | Descriptor: | 5'-D(*AP*GP*GP*AP*AP*TP*GP*GP*TP*CP*A)-3', 5'-D(*GP*AP*CP*CP*AP*TP*47C*CP*CP*T)-3', DNA POLYMERASE I, ... | | Authors: | Muenzel, M, Lercher, L, Mueller, M, Carell, T. | | Deposit date: | 2010-08-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Chemical Discrimination between Dc and 5Medc Via Their Hydroxylamine Adducts
Nucleic Acids Res., 38, 2010
|
|
3OJU
 
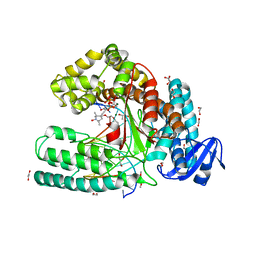 | | Snapshot of the large fragment of DNA polymerase I from Thermus Aquaticus processing c5 modified thymidies | | Descriptor: | 2'-deoxy-5-[(1-hydroxy-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)ethynyl]uridine 5'-(tetrahydrogen triphosphate), DNA (5'-D(*AP*AP*AP*AP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2010-08-23 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the synthesis of nucleobase modified DNA by Thermus aquaticus DNA polymerase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OJS
 
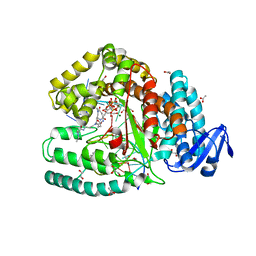 | | Snapshots of the large fragment of DNA polymerase I from Thermus Aquaticus processing C5 modified thymidines | | Descriptor: | 2'-deoxy-5-[9-(3-{[4-(diethylamino)-4-oxobutanoyl]amino}propyl)-18-ethyl-5,8,14,17-tetraoxo-4,9,13,18-tetraazaicos-1-yn-1-yl]uridine 5'-(tetrahydrogen triphosphate), ACETATE ION, DNA (5'-D(*AP*AP*AP*AP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2010-08-23 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the synthesis of nucleobase modified DNA by Thermus aquaticus DNA polymerase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XY7
 
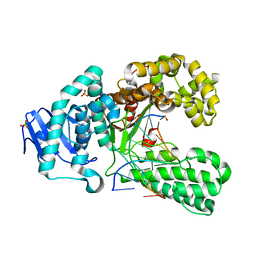 | | Crystal structure of a salicylic aldehyde base in the pre-insertion site of fragment DNA polymerase I from Bacillus stearothermophilus | | Descriptor: | 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*GP)-3', 5'-D(*SAYP*CP*GP*AP*GP*TP*CP*AP*GP*GP*CP)-3', DNA POLYMERASE I, ... | | Authors: | Kaul, C, Mueller, M, Wagner, M, Schneider, S, Carell, T. | | Deposit date: | 2010-11-15 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Reversible Bond Formation Enables the Replication and Amplification of a Crosslinking Salen Complex as an Orthogonal Base Pair.
Nature Chem., 3, 2011
|
|
2XY5
 
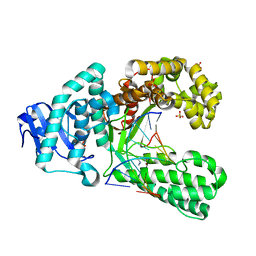 | | Crystal structure of an artificial salen-copper basepair in complex with fragment DNA polymerase I from Bacillus stearothermophilus | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 5'-D(*AP*GP*GP*GP*AP*SAYP*GP*GP*TP*CP)-3', 5'-D(*GP*AP*CP*CP*SAYP*TP*CP*CP*CP*TP)-3', ... | | Authors: | Kaul, C, Mueller, M, Wagner, M, Schneider, S, Carell, T. | | Deposit date: | 2010-11-15 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Reversible Bond Formation Enables the Replication and Amplification of a Crosslinking Salen Complex as an Orthogonal Base Pair.
Nature Chem., 3, 2011
|
|
2XY6
 
 | | Crystal structure of a salicylic aldehyde basepair in complex with fragment DNA polymerase I from Bacillus stearothermophilus | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*AP*GP*GP*GP*AP*SAYP*GP*GP*TP*CP)-3', ... | | Authors: | Kaul, C, Mueller, M, Wagner, M, Schneider, S, Carell, T. | | Deposit date: | 2010-11-15 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reversible Bond Formation Enables the Replication and Amplification of a Crosslinking Salen Complex as an Orthogonal Base Pair.
Nature Chem., 3, 2011
|
|
