4B9N
 
 | | Structure of the high fidelity DNA polymerase I correctly bypassing the oxidative formamidopyrimidine-dA DNA lesion. | | Descriptor: | 5'-D(*CP*AP*AP*(FAX)*AP*GP*AP*GP*TP*CP*AP*GP*GP*CP*TP)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*TP*TP*TP*TP)-3', DNA POLYMERASE, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4B9L
 
 | | Structure of the high fidelity DNA polymerase I with the oxidative formamidopyrimidine-dA DNA lesion in the pre-insertion site. | | Descriptor: | 5'-D(*CP*AP*GP*FAX*AP*GP*AP*GP*TP*CP*AP*GP*GP*CP*TP)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*TP)-3', DNA POLYMERASE I, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4B9S
 
 | | Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dG DNA lesion outside of the pre-insertion site. | | Descriptor: | 5'-D(*AP*CP*CP*TP*GP*AP*CP*TP*CP*TP)-3', 5'-D(*CP*AP*TP*FOXP*AP*GP*AP*GP*TP*CP*AP*GP*GP*TP*TP)-3', DNA POLYMERASE, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4BDP
 
 | | CRYSTAL STRUCTURE OF BACILLUS DNA POLYMERASE I FRAGMENT COMPLEXED TO 11 BASE PAIRS OF DUPLEX DNA AFTER ADDITION OF TWO DATP RESIDUES | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*AP*TP*GP*CP*AP*A)-3'), DNA (5'-D(*TP*AP*TP*TP*GP*CP*AP*TP*GP*AP*TP*GP*C)-3'), MAGNESIUM ION, ... | | Authors: | Kiefer, J.R, Mao, C, Beese, L.S. | | Deposit date: | 1997-11-17 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal.
Nature, 391, 1998
|
|
4B9T
 
 | | Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dG DNA lesion -dC basepair in the post-insertion site. | | Descriptor: | 5'-D(*AP*CP*CP*TP*GP*AP*CP*TP*CP*TP)-3', 5'-D(*CP*AP*TP*FOXP*AP*GP*AP*GP*TP*CP*AP*GP*GP*TP*TP)-3', DNA POLYMERASE, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4B9M
 
 | | Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dA DNA lesion -thymine basepair in the post- insertion site. | | Descriptor: | 5'-D(*DC*DA*DA*FAX*AP*GP*AP*GP*TP*CP*AP*GP*GP*CP*TP)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*TP*TP)-3', DNA POLYMERASE I, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4B9U
 
 | | Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dG DNA lesion -dA basepair in the post-insertion site. | | Descriptor: | 5'-D(*CP*AP*AP*FOXP*CP*GP*AP*GP*TP*CP*AP*GP*GP*CP*TP)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*GP*AP)-3', DNA POLYMERASE, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4B9V
 
 | | Structure of the high fidelity DNA polymerase I with extending from an oxidative formamidopyrimidine-dG DNA lesion -dA basepair. | | Descriptor: | 5'-D(*CP*AP*TP*FOXP*AP*GP*AP*GP*TP*CP*AP*GP*GP*CP*TP)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*TP*AP*AP)-3', DNA POLYMERASE, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
2HHU
 
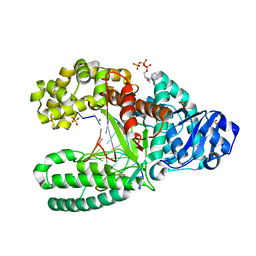 | | C:O6-methyl-guanine in the polymerase postinsertion site (-1 basepair position) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*GP*CP*GP*AP*TP*CP*AP*GP*CP*CP*C)-3', 5'-D(*GP*TP*AP*CP*(6OG)P*GP*GP*CP*TP*GP*AP*TP*CP*GP*CP*A)-3', ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HHV
 
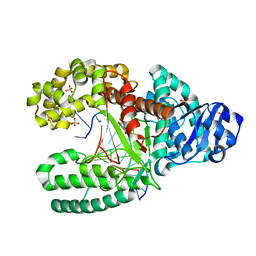 | | T:O6-methyl-guanine in the polymerase-2 basepair position | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*AP*GP*CP*TP*TP*G)-3', 5'-D(*GP*TP*AP*CP*(6OG)P*AP*GP*CP*TP*GP*AP*TP*CP*GP*CP*A)-3', DNA Polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HHX
 
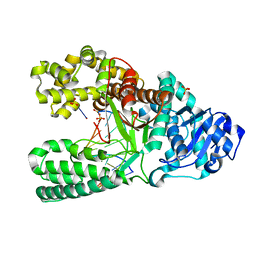 | | O6-methyl-guanine in the polymerase template preinsertion site | | Descriptor: | 5'-D(*CP*AP*TP*(6OG)P*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*G)-3', DNA Polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HVH
 
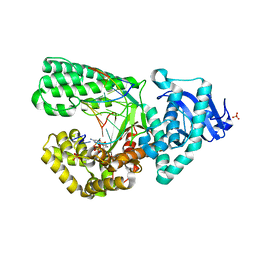 | | ddCTP:O6MeG pair in the polymerase active site (0 position) | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*CP*A*TP*(6OG)P*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*(DDG))-3', ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-07-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HVI
 
 | | ddCTP:G pair in the polymerase active site (0 position) | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*C*AP*TP*GP*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*(DDG))-3', ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-07-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HW3
 
 | | T:O6-methyl-guanine pair in the polymerase postinsertion site (-1 basepair position) | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*AP*GP*CP*TP*T)-3', 5'-D(*GP*TP*A*CP*(6OG)P*AP*GP*CP*TP*GP*AP*TP*CP*GP*CP*A)-3', DNA Polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-07-31 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6DSV
 
 | | Bst DNA polymerase I post-chemistry (n+2) structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), DNA polymerase I, ... | | Authors: | Chim, N, Jackson, L.N, Chaput, J.C. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Elife, 7, 2018
|
|
6DSW
 
 | | Bst DNA polymerase I pre-chemistry (n) structure | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*G)-3'), DNA (5'-D(P*AP*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Chim, N, Jackson, L.N, Chaput, J.C. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Elife, 7, 2018
|
|
6DSY
 
 | | Bst DNA polymerase I post-chemistry (n+1) structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3'), DNA (5'-D(P*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), DNA polymerase I, ... | | Authors: | Chim, N, Jackson, L.N, Chaput, J.C. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Elife, 7, 2018
|
|
6DSU
 
 | | Bst DNA polymerase I pre-insertion complex structure | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Chim, N, Jackson, L.N, Chaput, J.C. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Elife, 7, 2018
|
|
6DSX
 
 | | Bst DNA polymerase I post-chemistry (n+1 with dATP soak) structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), DNA polymerase I, ... | | Authors: | Chim, N, Jackson, L.N, Chaput, J.C. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Elife, 7, 2018
|
|
8SCN
 
 | | Bst DNA polymerase I Large Fragment mutant F710Y/D598A with 3'-amino primer, dGTP, and calcium time-resolved 24h (Product State) | | Descriptor: | CALCIUM ION, DIPHOSPHATE, DNA polymerase I, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCJ
 
 | | Bst DNA polymerase I Large Fragment mutant F710Y/D598A with 3'-amino primer, dGTP, and calcium time-resolved 2h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCS
 
 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 8h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCU
 
 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 48h (Product State) | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCI
 
 | | Bst DNA polymerase I Large Fragment mutant F710Y/D598A with 3'-amino primer, dGTP, and calcium time-resolved 1h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCM
 
 | | Bst DNA polymerase I Large Fragment mutant F710Y/D598A with 3'-amino primer, dGTP, and calcium time-resolved 8h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
