5ZS8
 
 | |
5ZR2
 
 | |
6CNL
 
 | | Crystal Structure of H105A PGAM5 Dodecamer | | Descriptor: | MAGNESIUM ION, PGAM5 Multimerization Motif Peptide, Serine/threonine-protein phosphatase PGAM5, ... | | Authors: | Ruiz, K, Agnew, C, Jura, N. | | Deposit date: | 2018-03-08 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional role of PGAM5 multimeric assemblies and their polymerization into filaments.
Nat Commun, 10, 2019
|
|
6CNI
 
 | | Crystal structure of H105A PGAM5 dimer | | Descriptor: | PHOSPHATE ION, SODIUM ION, Serine/threonine-protein phosphatase PGAM5, ... | | Authors: | Ruiz, K, Agnew, C, Jura, N. | | Deposit date: | 2018-03-08 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional role of PGAM5 multimeric assemblies and their polymerization into filaments.
Nat Commun, 10, 2019
|
|
1FBT
 
 | | THE BISPHOSPHATASE DOMAIN OF THE BIFUNCTIONAL RAT LIVER 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | FRUCTOSE-2,6-BISPHOSPHATASE, PHOSPHATE ION | | Authors: | Lee, Y.-H, Ogata, C, Pflugrath, J.W, Levitt, D.G, Sarma, R, Banaszak, L.J, Pilkis, S.J. | | Deposit date: | 1996-03-08 | | Release date: | 1997-07-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the rat liver fructose-2,6-bisphosphatase based on selenomethionine multiwavelength anomalous dispersion phases.
Biochemistry, 35, 1996
|
|
1E58
 
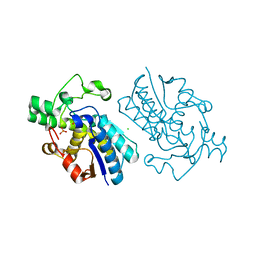 | | E.coli cofactor-dependent phosphoglycerate mutase | | Descriptor: | CHLORIDE ION, PHOSPHOGLYCERATE MUTASE, SULFATE ION | | Authors: | Bond, C.S, Hunter, W.N. | | Deposit date: | 2000-07-19 | | Release date: | 2001-03-20 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High Resolution Structure of the Phosphohistidine-Activated Form of Escherichia Coli Cofactor-Dependent Phosphoglycerate Mutase.
J.Biol.Chem., 276, 2001
|
|
1FZT
 
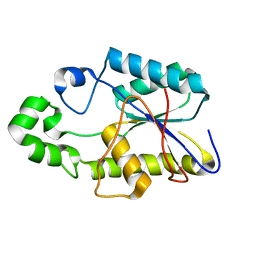 | | SOLUTION STRUCTURE AND DYNAMICS OF AN OPEN B-SHEET, GLYCOLYTIC ENZYME-MONOMERIC 23.7 KDA PHOSPHOGLYCERATE MUTASE FROM SCHIZOSACCHAROMYCES POMBE | | Descriptor: | PHOSPHOGLYCERATE MUTASE | | Authors: | Uhrinova, S, Uhrin, D, Nairn, J, Price, N.C, Fothergill-Gilmore, L.A. | | Deposit date: | 2000-10-04 | | Release date: | 2001-03-14 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of an open beta-sheet, glycolytic enzyme, monomeric 23.7 kDa phosphoglycerate mutase from Schizosaccharomyces pombe.
J.Mol.Biol., 306, 2001
|
|
1EBB
 
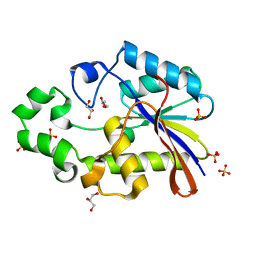 | | Bacillus stearothermophilus YhfR | | Descriptor: | GLYCEROL, PHOSPHATASE, SULFATE ION | | Authors: | Rigden, D.J, Jedrzejas, M.J. | | Deposit date: | 2001-07-24 | | Release date: | 2002-02-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of Action of a Cofactor-Dependent Phosphoglycerate Mutase Homolog from Bacillus Stearothermophilus with Broad Specificity Phosphatase Activity.
J.Mol.Biol., 315, 2002
|
|
1E59
 
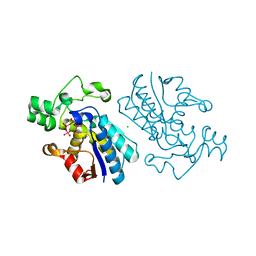 | |
1C81
 
 | | MICHAELIS COMPLEX OF FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1-deoxy-1-phosphono-6-O-phosphono-D-glucitol, FRUCTOSE-2,6-BISPHOSPHATASE | | Authors: | Lee, Y.H, Olson, T.W, McClard, R.W, Witte, J.F, McFarlan, S.C, Banaszak, L.J, Levitt, D.G, Lange, A.J. | | Deposit date: | 2000-04-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reaction Mechanism of Fructose-2,6-bisphosphatase Suggested by the Crystal Structures of a pseudo-Michaelis complex and Metabolite Complexes
To be Published
|
|
1H2F
 
 | |
2H52
 
 | |
3LNT
 
 | |
3LL4
 
 | | Structure of the H13A mutant of Ykr043C in complex with fructose-1,6-bisphosphate | | Descriptor: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), Uncharacterized protein YKR043C | | Authors: | Singer, A, Xu, X, Cui, H, Dong, A, Stogios, P.J, Edwards, A.M, Joachimiak, A, Savchenko, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure and activity of the metal-independent fructose-1,6-bisphosphatase YK23 from Saccharomyces cerevisiae.
J.Biol.Chem., 285, 2010
|
|
1QHF
 
 | | YEAST PHOSPHOGLYCERATE MUTASE-3PG COMPLEX STRUCTURE TO 1.7 A | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, PROTEIN (PHOSPHOGLYCERATE MUTASE), SULFATE ION | | Authors: | Crowhurst, G, Littlechild, J, Watson, H.C. | | Deposit date: | 1999-05-13 | | Release date: | 1999-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a phosphoglycerate mutase:3-phosphoglyceric acid complex at 1.7 A.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2HHJ
 
 | | Human bisphosphoglycerate mutase complexed with 2,3-bisphosphoglycerate (15 days) | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, 3-PHOSPHOGLYCERIC ACID, Bisphosphoglycerate mutase, ... | | Authors: | Wang, Y, Gong, W. | | Deposit date: | 2006-06-28 | | Release date: | 2006-10-24 | | Last modified: | 2016-07-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Seeing the process of histidine phosphorylation in human bisphosphoglycerate mutase.
J.Biol.Chem., 281, 2006
|
|
2HIA
 
 | |
3LG2
 
 | | A Ykr043C/ fructose-1,6-bisphosphate product complex following ligand soaking | | Descriptor: | PHOSPHATE ION, Uncharacterized protein YKR043C | | Authors: | Singer, A, Xu, X, Cui, H, Dong, A, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-19 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and activity of the metal-independent fructose-1,6-bisphosphatase YK23 from Saccharomyces cerevisiae.
J.Biol.Chem., 285, 2010
|
|
1RII
 
 | | Crystal structure of phosphoglycerate mutase from M. Tuberculosis | | Descriptor: | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase, GLYCEROL | | Authors: | Mueller, P, Sawaya, M.R, Chan, S, Wu, Y, Pashkova, I, Perry, J, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-11-17 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.70 angstroms X-ray crystal structure of Mycobacterium tuberculosis phosphoglycerate mutase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2IKQ
 
 | |
3NFY
 
 | |
3O0T
 
 | | Crystal structure of human phosphoglycerate mutase family member 5 (PGAM5) in complex with phosphate | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Serine/threonine-protein phosphatase PGAM5, ... | | Authors: | Chaikuad, A, Alfano, I, Picaud, S, Filippakopoulos, P, Barr, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Takeda, K, Ichijo, H, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-20 | | Release date: | 2010-10-06 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of PGAM5 Provide Insight into Active Site Plasticity and Multimeric Assembly.
Structure, 25, 2017
|
|
3MBK
 
 | |
8IT7
 
 | | Phosphoglycerate mutase 1 complexed with a compound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(4-chlorophenyl)-2-oxidanyl-6-[(4-phenylphenyl)sulfonylamino]benzoic acid, CHLORIDE ION, ... | | Authors: | Zhou, L, Jiang, L.L. | | Deposit date: | 2023-03-22 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phosphoglycerate mutase 1 complexed with a compound
To Be Published
|
|
8ITD
 
 | | Phosphoglycerate mutase 1 complexed with a compound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(4-chloranyl-2-oxidanyl-phenyl)-2-[(4-phenylphenyl)sulfonylamino]benzoic acid, CHLORIDE ION, ... | | Authors: | Zhou, L, Jiang, L.L. | | Deposit date: | 2023-03-22 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phosphoglycerate mutase 1 complexed with a compound
To Be Published
|
|
