1W93
 
 | | Crystal Structure of Biotin Carboxylase Domain of Acetyl-Coenzyme A Carboxylase from Saccharomyces cerevisiae | | Descriptor: | ACETYL-COENZYME A CARBOXYLASE | | Authors: | Shen, Y, Volrath, S.L, Weatherly, S.C, Elich, T.D, Tong, L. | | Deposit date: | 2004-10-05 | | Release date: | 2005-01-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Mechanism for the Potent Inhibition of Eukaryotic Acetyl-Coenzyme a Carboxylase by Soraphen A, a Macrocyclic Polyketide Natural Product
Mol.Cell, 16, 2004
|
|
5CSL
 
 | |
5KKN
 
 | | Crystal structure of human ACC2 BC domain in complex with ND-646, the primary amide of ND-630 | | Descriptor: | 2-[1-[(2~{R})-2-(2-methoxyphenyl)-2-(oxan-4-yloxy)ethyl]-5-methyl-6-(1,3-oxazol-2-yl)-2,4-bis(oxidanylidene)thieno[2,3-d]pyrimidin-3-yl]-2-methyl-propanamide, Acetyl-CoA carboxylase 2 | | Authors: | Wang, R, Paul, D, Tong, L. | | Deposit date: | 2016-06-22 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Acetyl-CoA carboxylase inhibition by ND-630 reduces hepatic steatosis, improves insulin sensitivity, and modulates dyslipidemia in rats.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1W96
 
 | | Crystal Structure of Biotin Carboxylase Domain of Acetyl-coenzyme A Carboxylase from Saccharomyces cerevisiae in Complex with Soraphen A | | Descriptor: | ACETYL-COENZYME A CARBOXYLASE, SORAPHEN A | | Authors: | Shen, Y, Volrath, S.L, Weatherly, S.C, Elich, T.D, Tong, L. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Mechanism for the Potent Inhibition of Eukaryotic Acetyl-Coenzyme a Carboxylase by Soraphen A, a Macrocyclic Polyketide Natural Product
Mol.Cell, 16, 2004
|
|
1BNC
 
 | |
3N6R
 
 | | CRYSTAL STRUCTURE OF the holoenzyme of PROPIONYL-COA CARBOXYLASE (PCC) | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Propionyl-CoA carboxylase, alpha subunit, ... | | Authors: | Huang, C.S, Sadre-Bazzaz, K, Tong, L. | | Deposit date: | 2010-05-26 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the alpha(6)beta(6) holoenzyme of propionyl-coenzyme A carboxylase.
Nature, 466, 2010
|
|
1ULZ
 
 | | Crystal structure of the biotin carboxylase subunit of pyruvate carboxylase | | Descriptor: | pyruvate carboxylase n-terminal domain | | Authors: | Kondo, S, Nakajima, Y, Sugio, S, Yong-Biao, J, Sueda, S, Kondo, H. | | Deposit date: | 2003-09-18 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the biotin carboxylase subunit of pyruvate carboxylase from Aquifex aeolicus at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5CSK
 
 | |
2YL2
 
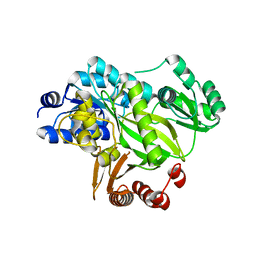 | | Crystal structure of human acetyl-CoA carboxylase 1, biotin carboxylase (BC) domain | | Descriptor: | ACETYL-COA CARBOXYLASE 1 | | Authors: | Muniz, J.R.C, Froese, D.S, Krysztofinska, E, Vollmar, M, Beltrami, A, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Yue, W.W, Oppermann, U. | | Deposit date: | 2011-05-31 | | Release date: | 2011-06-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Human Acetyl-Coa Carboxylase 1, Biotin Carboxylase (Bc) Domain
To be Published
|
|
5KS8
 
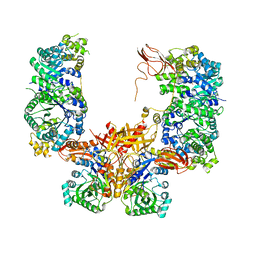 | |
1DV1
 
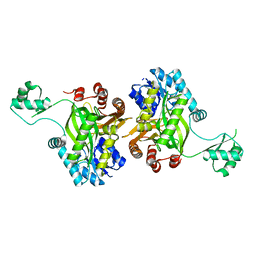 | | STRUCTURE OF BIOTIN CARBOXYLASE (APO) | | Descriptor: | BIOTIN CARBOXYLASE, PHOSPHATE ION | | Authors: | Thoden, J.B, Blanchard, C.Z, Holden, H.M, Waldrop, G.L. | | Deposit date: | 2000-01-19 | | Release date: | 2000-06-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Movement of the biotin carboxylase B-domain as a result of ATP binding.
J.Biol.Chem., 275, 2000
|
|
3TW6
 
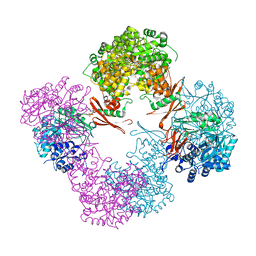 | | Structure of Rhizobium etli pyruvate carboxylase T882A with the allosteric activator, acetyl coenzyme-A | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | St Maurice, M, Kumar, S, Lietzan, A.D. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-19 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Interaction between the biotin carboxyl carrier domain and the biotin carboxylase domain in pyruvate carboxylase from Rhizobium etli.
Biochemistry, 50, 2011
|
|
2C00
 
 | |
3TW7
 
 | | Structure of Rhizobium etli pyruvate carboxylase T882A crystallized without acetyl coenzyme-A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Pyruvate carboxylase protein, ... | | Authors: | St Maurice, M, Kumar, S, Lietzan, A.D. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-12 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Interaction between the biotin carboxyl carrier domain and the biotin carboxylase domain in pyruvate carboxylase from Rhizobium etli.
Biochemistry, 50, 2011
|
|
1DV2
 
 | | The structure of biotin carboxylase, mutant E288K, complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, BIOTIN CARBOXYLASE | | Authors: | Thoden, J.B, Blanchard, C.Z, Holden, H.M, Waldrop, G.L. | | Deposit date: | 2000-01-19 | | Release date: | 2000-06-09 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Movement of the biotin carboxylase B-domain as a result of ATP binding.
J.Biol.Chem., 275, 2000
|
|
8F41
 
 | | 3-methylcrotonyl-CoA carboxylase in filament, alpha-subunit centered | | Descriptor: | 3-methylcrotonyl-CoA carboxylase, alpha-subunit, beta-subunit, ... | | Authors: | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-11-10 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
8F3D
 
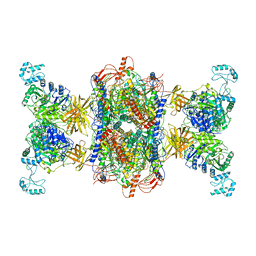 | | 3-methylcrotonyl-CoA carboxylase in filament, beta-subunit centered | | Descriptor: | 3-methylcrotonyl-CoA carboxylase alpha-subunit, 3-methylcrotonyl-CoA carboxylase beta-subunit, 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL | | Authors: | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-11-09 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
6YBP
 
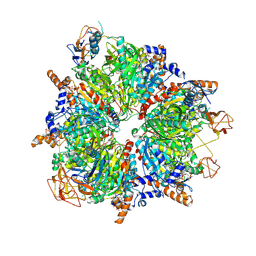 | | Propionyl-CoA carboxylase of Methylorubrum extorquens with bound CoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Propionyl-CoA carboxylase alpha subunit, ... | | Authors: | Schuller, J.M, Schuller, S.K, Zarzycki, J, Scheffen, M, Marchal, D.M, Erb, T.J. | | Deposit date: | 2020-03-17 | | Release date: | 2020-10-28 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation
Nat Catal, 2021
|
|
5MLK
 
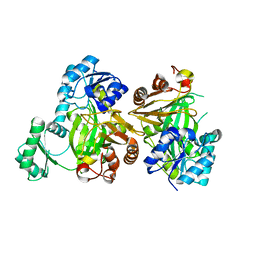 | |
6YBQ
 
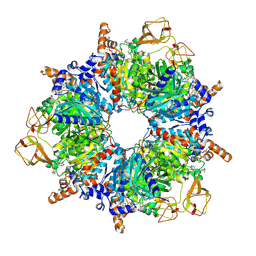 | | Engineered glycolyl-CoA carboxylase (quintuple mutant) with bound CoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Propionyl-CoA carboxylase alpha subunit, ... | | Authors: | Schuller, J.M, Schuller, S.K, Zarzycki, J, Scheffen, M, Marchal, D.M, Erb, T.J. | | Deposit date: | 2020-03-17 | | Release date: | 2020-10-28 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (1.96 Å) | | Cite: | A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation
Nat Catal, 2021
|
|
4RCN
 
 | |
3BG5
 
 | | Crystal Structure of Staphylococcus Aureus Pyruvate Carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Xiang, S, Tong, L. | | Deposit date: | 2007-11-26 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of human and Staphylococcus aureus pyruvate carboxylase and molecular insights into the carboxyltransfer reaction.
Nat.Struct.Mol.Biol., 15, 2008
|
|
4RZQ
 
 | | Structural Analysis of Substrate, Reaction Intermediate and Product Binding in Haemophilus influenzae Biotin Carboxylase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Biotin carboxylase, methyl (3aS,4S,6aR)-4-(5-methoxy-5-oxopentyl)-2-oxohexahydro-1H-thieno[3,4-d]imidazole-1-carboxylate | | Authors: | Broussard, T.C, Pakhomova, S, Neau, D.B, Champion, T.S, Bonnot, R, Waldrop, G.L. | | Deposit date: | 2014-12-23 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry, 54, 2015
|
|
2DZD
 
 | | Crystal structure of the biotin carboxylase domain of pyruvate carboxylase | | Descriptor: | pyruvate carboxylase | | Authors: | Kondo, S, Nakajima, Y, Sugio, S, Sueda, S, Islam, M.N, Kondo, H. | | Deposit date: | 2006-09-27 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the biotin carboxylase domain of pyruvate carboxylase from Bacillus thermodenitrificans
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
3U9T
 
 | |
