7TNY
 
 | | Cryo-EM structure of RIG-I in complex with p2dsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, ZINC ION, p2dsRNA | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
4C9B
 
 | | Crystal structure of eIF4AIII-CWC22 complex | | Descriptor: | EUKARYOTIC INITIATION FACTOR 4A-III, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Buchwald, G, Schuessler, S, Basquin, C, LeHir, H, Conti, E. | | Deposit date: | 2013-10-02 | | Release date: | 2013-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human Eif4Aiii-Cwc22 Complex Shows How a Dead-Box Protein is Inhibited by a Mif4G Domain
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4CDG
 
 | | Crystal structure of the Bloom's syndrome helicase BLM in complex with Nanobody | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BLOOM SYNDROME PROTEIN, NANOBODY, ... | | Authors: | Newman, J.A, Savitsky, P, Allerston, C.K, Pike, A.C.W, Pardon, E, Steyaert, J, Arrowsmith, C.H, von Delft, F, Bountra, C, Edwards, A, Gileadi, O. | | Deposit date: | 2013-10-31 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal Structure of the Bloom'S Syndrome Helicase Indicates a Role for the Hrdc Domain in Conformational Changes.
Nucleic Acids Res., 43, 2015
|
|
4BPB
 
 | | STRUCTURAL INSIGHTS INTO RNA RECOGNITION BY RIG-I | | Descriptor: | 5'-R(*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP)-3', PROBABLE ATP-DEPENDENT RNA HELICASE DDX58, SULFATE ION, ... | | Authors: | Luo, D, Pyle, A.M. | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.584 Å) | | Cite: | Structural Insights Into RNA Recognition by Rig-I.
Cell(Cambridge,Mass.), 147, 2011
|
|
4BUJ
 
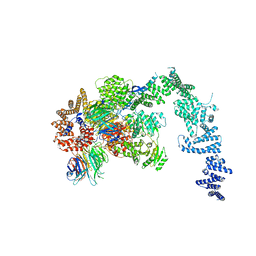 | | Crystal structure of the S. cerevisiae Ski2-3-8 complex | | Descriptor: | ANTIVIRAL HELICASE SKI2, ANTIVIRAL PROTEIN SKI8, SULFATE ION, ... | | Authors: | Halbach, F, Reichelt, P, Rode, M, Conti, E. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The Yeast Ski Complex: Crystal Structure and RNA Channeling to the Exosome Complex.
Cell(Cambridge,Mass.), 154, 2013
|
|
4BRW
 
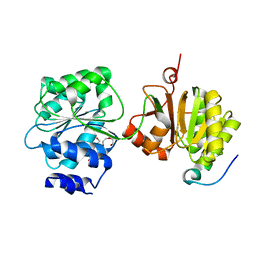 | | Crystal structure of the yeast Dhh1-Pat1 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-DEPENDENT RNA HELICASE DHH1, DNA TOPOISOMERASE 2-ASSOCIATED PROTEIN PAT1, ... | | Authors: | Sharif, H, Ozgur, S, Sharma, K, Basquin, C, Urlaub, H, Conti, E. | | Deposit date: | 2013-06-05 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | Structural Analysis of the Yeast Dhh1-Pat1 Complex Reveals How Dhh1 Engages Pat1, Edc3 and RNA in Mutually Exclusive Interactions
Nucleic Acids Res., 41, 2013
|
|
4BRU
 
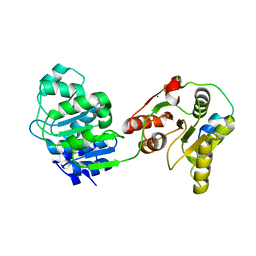 | | Crystal structure of the yeast Dhh1-Edc3 complex | | Descriptor: | ATP-DEPENDENT RNA HELICASE DHH1, ENHANCER OF MRNA-DECAPPING PROTEIN 3 | | Authors: | Sharif, H, Ozgur, S, Sharma, K, Basquin, C, Urlaub, H, Conti, E. | | Deposit date: | 2013-06-05 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.245 Å) | | Cite: | Structural Analysis of the Yeast Dhh1-Pat1 Complex Reveals How Dhh1 Engages Pat1, Edc3 and RNA in Mutually Exclusive Interactions
Nucleic Acids Res., 41, 2013
|
|
4CGZ
 
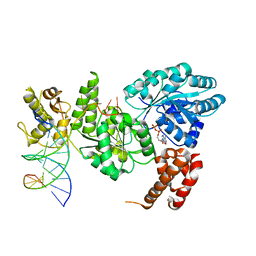 | | Crystal structure of the Bloom's syndrome helicase BLM in complex with DNA | | Descriptor: | 5'-D(*AP*GP*CP*GP*TP*CP*GP*AP*GP*AP*TP*CP)-3', 5'-D(*GP*AP*TP*CP*TP*CP*GP*AP*CP*GP*CP*TP*CP*DT *CP*CP*CP)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Newman, J.A, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2013-11-27 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the Bloom'S Syndrome Helicase Indicates a Role for the Hrdc Domain in Conformational Changes.
Nucleic Acids Res., 43, 2015
|
|
4CT4
 
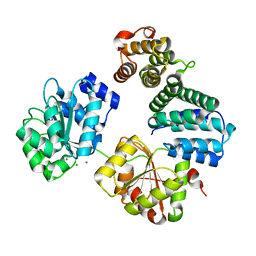 | | CNOT1 MIF4G domain - DDX6 complex | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ozgur, S, Basquin, J, Conti, E. | | Deposit date: | 2014-03-12 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Insights to the Role of the Ccr4- not Complex and Ddx6 ATPase in Microrna Repression.
Mol.Cell, 54, 2014
|
|
4D26
 
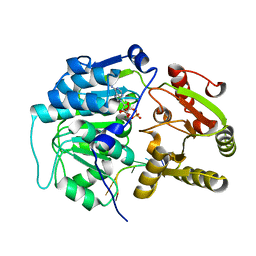 | | Crystal structure of the Bombyx mori Vasa helicase (E339Q) in complex with RNA,ADP and Pi | | Descriptor: | 5'-R(*UP*GP*AP*CP*AP*UP)-3', ADENOSINE-5'-DIPHOSPHATE, BMVLG PROTEIN, ... | | Authors: | Spinelli, P, Pillai, R.S, Kadlec, J, Cusack, S. | | Deposit date: | 2014-05-07 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | RNA Clamping by Vasa Assembles a Pirna Amplifier Complex on Transposon Transcripts.
Cell(Cambridge,Mass.), 157, 2014
|
|
6QIE
 
 | | Crystal structure of DEAH-box ATPase Prp43-S387G | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Hamann, F, Ficner, R, Enders, M. | | Deposit date: | 2019-01-18 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for RNA translocation by DEAH-box ATPases.
Nucleic Acids Res., 47, 2019
|
|
4D25
 
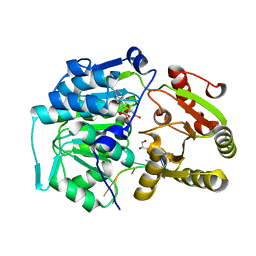 | | Crystal structure of the Bombyx mori Vasa helicase (E339Q) in complex with RNA and AMPPNP | | Descriptor: | 5'-R(*UP*GP*AP*CP*AP*UP)-3', BMVLG PROTEIN, GLYCEROL, ... | | Authors: | Spinelli, P, Pillai, R.S, Kadlec, J, Cusack, S. | | Deposit date: | 2014-05-07 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RNA Clamping by Vasa Assembles a Pirna Amplifier Complex on Transposon Transcripts.
Cell(Cambridge,Mass.), 157, 2014
|
|
4CT5
 
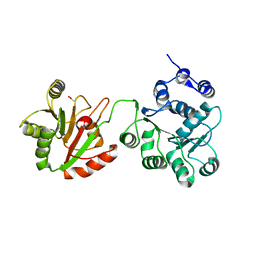 | | DDX6 | | Descriptor: | ACETATE ION, DDX6 | | Authors: | Ozgur, S, Basquin, J, Conti, E. | | Deposit date: | 2014-03-12 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Biochemical Insights to the Role of the Ccr4-not Complex and Ddx6 ATPase in Microrna Repression.
Mol.Cell, 54, 2014
|
|
6QIC
 
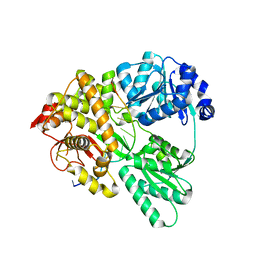 | |
4DDX
 
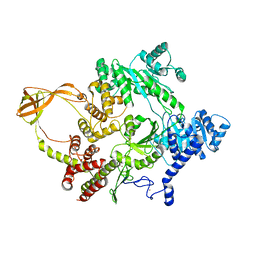 | |
6QID
 
 | | Crystal structure of DEAH-box ATPase Prp43-S387A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Hamann, F, Ficner, R, Enders, M. | | Deposit date: | 2019-01-18 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural basis for RNA translocation by DEAH-box ATPases.
Nucleic Acids Res., 47, 2019
|
|
4DDU
 
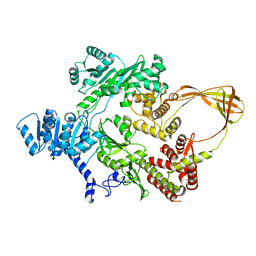 | | Thermotoga maritima reverse gyrase, C2 FORM 1 | | Descriptor: | Reverse gyrase, ZINC ION | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2012-01-19 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Thermotoga maritima reverse gyrase: inferences for the mechanism of positive DNA supercoiling.
Nucleic Acids Res., 41, 2013
|
|
4DDT
 
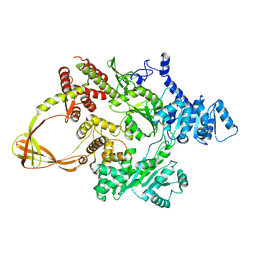 | | Thermotoga maritima reverse gyrase, C2 FORM 2 | | Descriptor: | Reverse gyrase, ZINC ION | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2012-01-19 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of Thermotoga maritima reverse gyrase: inferences for the mechanism of positive DNA supercoiling.
Nucleic Acids Res., 41, 2013
|
|
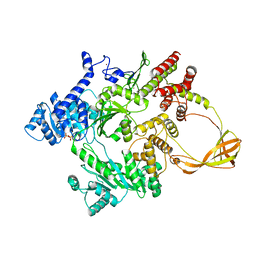
4DDW
 
 | |
6QWS
 
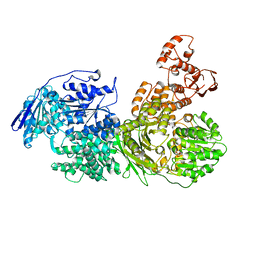 | |
6QV3
 
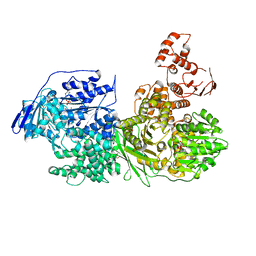 | | Crystal structure of the Ski2 RNA-helicase Brr2 from Chaetomium thermophilum bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Absmeier, E, Santos, K.F, Wahl, M.C. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Mechanism Underlying Inhibition of Intrinsic ATPase Activity in a Ski2-like RNA Helicase.
Structure, 28, 2020
|
|
4DDV
 
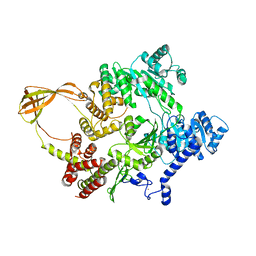 | |
6QV4
 
 | | Crystal structure of the Ski2 RNA-helicase Brr2 from Chaetomium thermophilum bound to ATP-gamma-S | | Descriptor: | ACETATE ION, MANGANESE (II) ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Absmeier, E, Santos, K.F, Wahl, M.C. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Mechanism Underlying Inhibition of Intrinsic ATPase Activity in a Ski2-like RNA Helicase.
Structure, 28, 2020
|
|
6RM9
 
 | | Crystal structure of the DEAH-box ATPase Prp2 in complex with Spp2 and ADP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2019-05-06 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6RMB
 
 | | Crystal structure of the DEAH-box ATPase Prp2 in complex with Spp2 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative mRNA splicing factor, ... | | Authors: | Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2019-05-06 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
