6KQF
 
 | | Thermus thermophilus initial transcription complex comprising sigma A and 5'-OH RNA of 5 nt | | Descriptor: | DNA (5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G*)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, Y, Li, L, Ebright, R.H. | | Deposit date: | 2019-08-17 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4XLS
 
 | |
4XLR
 
 | |
4XLN
 
 | |
6L74
 
 | | Thermus thermophilus initial transcription complex comprising sigma A and 5'-triphosphate RNA of 2 nt | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, Y, Li, L, Ebright, R.H. | | Deposit date: | 2019-10-31 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4XLQ
 
 | |
4XSZ
 
 | | Crystal structure of CBR 9393 bound to Escherichia coli RNA polymerase holoenzyme | | Descriptor: | 4-[3-(4-fluorophenyl)-1H-pyrazol-4-yl]-N-[2-(piperazin-1-yl)ethyl]-2-(trifluoromethyl)aniline, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2015-01-22 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.683 Å) | | Cite: | CBR antimicrobials inhibit RNA polymerase via at least two bridge-helix cap-mediated effects on nucleotide addition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XSX
 
 | | Crystal structure of CBR 703 bound to Escherichia coli RNA polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2015-01-22 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.708 Å) | | Cite: | CBR antimicrobials inhibit RNA polymerase via at least two bridge-helix cap-mediated effects on nucleotide addition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XLP
 
 | |
6LTS
 
 | |
4XSY
 
 | | Crystal structure of CBR 9379 bound to Escherichia coli RNA polymerase holoenzyme | | Descriptor: | 3-{[(2,6-dichlorophenyl)carbamoyl]amino}-N-hydroxy-N'-phenyl-5-(trifluoromethyl)benzenecarboximidamide, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2015-01-22 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.007 Å) | | Cite: | CBR antimicrobials inhibit RNA polymerase via at least two bridge-helix cap-mediated effects on nucleotide addition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6M7J
 
 | | Mycobacterium tuberculosis RNAP with RbpA/us fork and Corallopyronin | | Descriptor: | DNA (26-MER), DNA (31-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2018-08-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding.
Nature, 565, 2019
|
|
4YLP
 
 | | E. coli Transcription Initiation Complex - 16-bp spacer and 5-nt RNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, Steitz, T.A. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Crystal Structures of the E. coli Transcription Initiation Complexes with a Complete Bubble.
Mol.Cell, 58, 2015
|
|
4YG2
 
 | | X-ray crystal structur of Escherichia coli RNA polymerase sigma70 holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | X-ray crystal structure of Escherichia coli RNA polymerase sigma70 holoenzyme
J. Biol. Chem., 288, 2013
|
|
4YLO
 
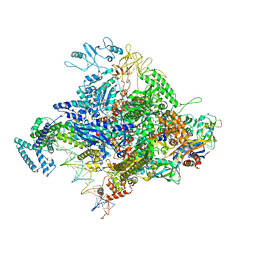 | | E. coli Transcription Initiation Complex - 16-bp spacer and 4-nt RNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, Steitz, T.A. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Crystal Structures of the E. coli Transcription Initiation Complexes with a Complete Bubble.
Mol.Cell, 58, 2015
|
|
4YLN
 
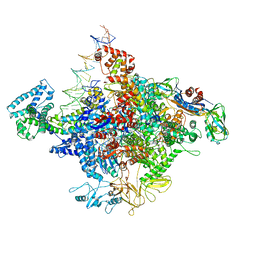 | | E. coli Transcription Initiation Complex - 17-bp spacer and 4-nt RNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, Steitz, T.A. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Crystal Structures of the E. coli Transcription Initiation Complexes with a Complete Bubble.
Mol.Cell, 58, 2015
|
|
4YFX
 
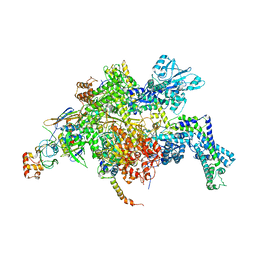 | | Escherichia coli RNA polymerase in complex with Myxopyronin B | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.844 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
6N62
 
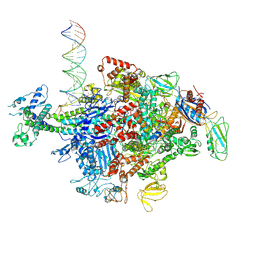 | | Escherichia coli RNA polymerase sigma70-holoenzyme bound to upstream fork promoter DNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Braffman, N, Hauver, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-24 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.803 Å) | | Cite: | Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4YFK
 
 | | Escherichia coli RNA polymerase in complex with squaramide compound 8. | | Descriptor: | 3,5-dimethyl-N-{2-[4-(4-methylbenzyl)piperidin-1-yl]-3,4-dioxocyclobut-1-en-1-yl}-1,2-oxazole-4-sulfonamide, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.571 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
6N4C
 
 | | EM structure of the DNA wrapping in bacterial open transcription initiation complex | | Descriptor: | DNA (94-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Florez-Ariza, A, Cassago, A, de Oliveira, P.S.L, Guerra, D.G, Portugal, R.V. | | Deposit date: | 2018-11-19 | | Release date: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Interactions of Upstream and Downstream Promoter Regions with RNA Polymerase are Energetically Coupled and a Target of Regulation in Transcription Initiation
Biorxiv, 2020
|
|
6N61
 
 | | Escherichia coli RNA polymerase sigma70-holoenzyme bound to upstream fork promoter DNA and Capistruin | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Capistruin, ... | | Authors: | Braffman, N, Hauver, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-24 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.253 Å) | | Cite: | Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4YFN
 
 | | Escherichia coli RNA polymerase in complex with squaramide compound 14 (N-[3,4-dioxo-2-(4-{[4-(trifluoromethyl)benzyl]amino}piperidin-1-yl)cyclobut-1-en-1-yl]-3,5-dimethyl-1,2-oxazole-4-sulfonamide) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.817 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
6N57
 
 | | Cryo-EM structure of Escherichia coli RNAP polymerase bound with TraR in conformation I | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-21 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | E. coliTraR allosterically regulates transcription initiation by altering RNA polymerase conformation.
Elife, 8, 2019
|
|
6N58
 
 | | Cryo-EM structure of Escherichia coli RNAP polymerase bound with TraR in conformation II | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-21 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | E. coliTraR allosterically regulates transcription initiation by altering RNA polymerase conformation.
Elife, 8, 2019
|
|
4ZH2
 
 | |
